+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Fab4251-DS-SOSIP complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HIV / VIRAL PROTEIN | |||||||||
| Biological species | HIV whole-genome vector AA1305#18 (others) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
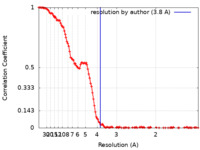
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Nortier P / Perez L | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Res Sq / Year: 2024 Journal: Res Sq / Year: 2024Title: RAIN: a Machine Learning-based identification for HIV-1 bNAbs. Authors: Laurent Perez / Mathilde Foglierini /  Abstract: Broadly neutralizing antibodies (bNAbs) are promising candidates for the treatment and prevention of HIV-1 infection. Despite their critical importance, automatic detection of HIV-1 bNAbs from immune ...Broadly neutralizing antibodies (bNAbs) are promising candidates for the treatment and prevention of HIV-1 infection. Despite their critical importance, automatic detection of HIV-1 bNAbs from immune repertoire is still lacking. Here, we developed a straightforward computational method for apid utomatic dentification of bAbs ) based on Machine Learning methods. In contrast to other approaches using one-hot encoding amino acid sequences or structural alignment for prediction, RAIN uses a combination of selected sequence-based features for accurate prediction of HIV-1 bNAbs. We demonstrate the performance of our approach on non-biased, experimentally obtained sequenced BCR repertoires from HIV-1 immune donors. RAIN processing leads to the successful identification of novel HIV-1 bNAbs targeting the CD4-binding site of the envelope glycoprotein. In addition, we validate the identified bNAbs using neutralization assay and we solve the structure of one of them in complex with the soluble native-like heterotrimeric envelope glycoprotein by single-particle cryo-electron microscopy (cryo-EM). Overall, we propose a method to facilitate and accelerate HIV-1 bNAbs discovery from non-selected immune repertoires. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19665.map.gz emd_19665.map.gz | 154.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19665-v30.xml emd-19665-v30.xml emd-19665.xml emd-19665.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19665_fsc.xml emd_19665_fsc.xml | 16.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_19665.png emd_19665.png | 28.9 KB | ||
| Filedesc metadata |  emd-19665.cif.gz emd-19665.cif.gz | 6.9 KB | ||
| Others |  emd_19665_half_map_1.map.gz emd_19665_half_map_1.map.gz emd_19665_half_map_2.map.gz emd_19665_half_map_2.map.gz | 151.8 MB 151.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19665 http://ftp.pdbj.org/pub/emdb/structures/EMD-19665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19665 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19665.map.gz / Format: CCP4 / Size: 198.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19665.map.gz / Format: CCP4 / Size: 198.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.73277 Å | ||||||||||||||||||||||||||||||||||||
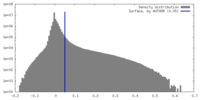
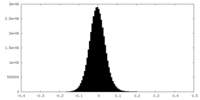
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_19665_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
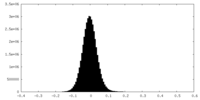
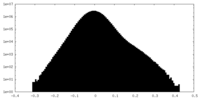
| Density Histograms |
-Half map: #2
| File | emd_19665_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
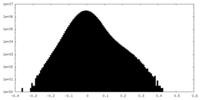
| Density Histograms |
- Sample components
Sample components
-Entire : Envelope glycoprotein Human immunodeficiency virus 1 with Fab
| Entire | Name: Envelope glycoprotein Human immunodeficiency virus 1 with Fab |
|---|---|
| Components |
|
-Supramolecule #1: Envelope glycoprotein Human immunodeficiency virus 1 with Fab
| Supramolecule | Name: Envelope glycoprotein Human immunodeficiency virus 1 with Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism: HIV whole-genome vector AA1305#18 (others) |
| Molecular weight | Theoretical: 400 kDa/nm |
-Macromolecule #1: variable heavy chain
| Macromolecule | Name: variable heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.951986 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVHVMQSGDQ VKKPGASVQV SCTTSGSSFI EDSLHWIQQV PGQEPEWLGW VNPRHGAVNY SWKIRDRITM TRDISKSTMY VQMRGLQSD DTAMYYCAKS RRGANWALYW GWGTRITVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK DYFPEPVTVS W NSGALTSG ...String: QVHVMQSGDQ VKKPGASVQV SCTTSGSSFI EDSLHWIQQV PGQEPEWLGW VNPRHGAVNY SWKIRDRITM TRDISKSTMY VQMRGLQSD DTAMYYCAKS RRGANWALYW GWGTRITVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK DYFPEPVTVS W NSGALTSG VHTFPAVLQS SGLYSLSSVV TVPSSSLGTQ TYICNVNHKP SNTKVDKKVE PK |
-Macromolecule #2: variable light chain
| Macromolecule | Name: variable light chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.257809 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQLTQSPSY LAASVGDRVT ITCRESNDLN WYLQRPGKPP KLLISGASKL ERGVPSRFRG SSSGSLTISG LQPEDIGTYY CQVFEFFGG GTRVDIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD S TYSLSSTL ...String: DIQLTQSPSY LAASVGDRVT ITCRESNDLN WYLQRPGKPP KLLISGASKL ERGVPSRFRG SSSGSLTISG LQPEDIGTYY CQVFEFFGG GTRVDIKRTV AAPSVFIFPP SDEQLKSGTA SVVCLLNNFY PREAKVQWKV DNALQSGNSQ ESVTEQDSKD S TYSLSSTL TLSKADYEKH KVYACEVTHQ GLSSPVTKSF NRGEC |
-Macromolecule #3: Envelope glycoprotein gp120
| Macromolecule | Name: Envelope glycoprotein gp120 / type: protein_or_peptide / ID: 3 Details: Envelope glycoprotein gp120|Human immunodeficiency virus 1 Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: HIV whole-genome vector AA1305#18 (others) |
| Molecular weight | Theoretical: 49.558484 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKDAET TLFCASDHNV WATHACVPTD PNPQEIHLEN VTEEFNMWKN NMVEQMHTDI ISLWDQSLKP CVKLTPLCV TLQCTNVTNN ITDDMRGELK NCSFNMTTEL RDKKQKVYSL FYRLDVVQIK EYRLINCNTS ACTQACPKVS F EPIPIHYC ...String: AENLWVTVYY GVPVWKDAET TLFCASDHNV WATHACVPTD PNPQEIHLEN VTEEFNMWKN NMVEQMHTDI ISLWDQSLKP CVKLTPLCV TLQCTNVTNN ITDDMRGELK NCSFNMTTEL RDKKQKVYSL FYRLDVVQIK EYRLINCNTS ACTQACPKVS F EPIPIHYC APAGFAILKC KDKKFNGTGP CPSVSTVQCT HGIKPVVSTQ LLLNGSLAEE EVMIRSENIT NNAKNILVQF NT PVQINCT RPNNNTRKSI RIGPGQAFYA TGDIIGDIRQ AHCNVSKATW NETLGKVVKQ LRKHFGNNTI IRFANSSGGD LEV TTHSFN CGGEFFYCNT SGLFNSTWIS NSNDSITLPC RIKQIINMWQ RIGQCMYAPP IQGVIRCVSN ITGLILTRDG GSTN STTET FRPGGGDMRD NWRSELYKYK VVKIEPLGVA PTRCKR |
-Macromolecule #4: Envelope glycoprotein gp41
| Macromolecule | Name: Envelope glycoprotein gp41 / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: HIV whole-genome vector AA1305#18 (others) |
| Molecular weight | Theoretical: 14.561529 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSTVWGI KQLQARVLAV ERYLRDQQLL GIWGCSGKLI CCTNVPWNSS WSNRNLSEI WDNMTWLQWD KEISNYTQII YGLLEESQNQ QEKNEQDLLA L |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 17 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 6 |
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 4 / Number real images: 15163 / Average electron dose: 39.89 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-8s2e: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)