[English] 日本語
 Yorodumi
Yorodumi- EMDB-19276: Cryo-EM of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleav... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) (C1) | ||||||||||||
 Map data Map data | Sharpened cryo-EM map of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) (C1) | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Cell adhesion / fertilization / egg-sperm interaction / gamete recognition / sperm receptor / extracellular matrix / egg coat / zona pellucida / glycoprotein / N-glycan / structural protein / ZP-N domain / block to polyspermy / post-fertilization cleavage / ovastacin | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationacrosin binding / structural constituent of egg coat / egg coat / prevention of polyspermy / binding of sperm to zona pellucida / extracellular region / plasma membrane Similarity search - Function | ||||||||||||
| Biological species | |||||||||||||
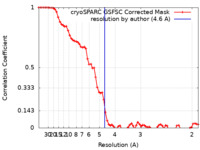
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | ||||||||||||
 Authors Authors | Wiseman B / Nishio S / Jovine L | ||||||||||||
| Funding support |  Sweden, 3 items Sweden, 3 items
| ||||||||||||
 Citation Citation | Journal: Int J Dev Biol / Year: 2008 Title: Anuran and pig egg zona pellucida glycoproteins in fertilization and early development. Authors: Jerry L Hedrick /  Abstract: The envelope surrounding the eggs of all animals has many biological functions in fertilization and development. This review focuses on the anuran egg envelope in terms of its biochemistry, ...The envelope surrounding the eggs of all animals has many biological functions in fertilization and development. This review focuses on the anuran egg envelope in terms of its biochemistry, biophysics, structural biology and function in sperm-egg interactions and early development (embryo hatching). Egg envelopes from Xenopus laevis are among the most studied of the anurans, and are the central theme of this review. Comparisons of Xenopus laevis envelopes with those of other anurans and with pig egg envelopes are also included. This article presents historical as well as contemporary comparative perspectives on molecular and cellular mechanisms of sperm-egg envelope binding, block to polyspermy, envelope hardening, and hatching. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19276.map.gz emd_19276.map.gz | 28.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19276-v30.xml emd-19276-v30.xml emd-19276.xml emd-19276.xml | 28 KB 28 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19276_fsc.xml emd_19276_fsc.xml | 6.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_19276.png emd_19276.png | 83.4 KB | ||
| Filedesc metadata |  emd-19276.cif.gz emd-19276.cif.gz | 6.7 KB | ||
| Others |  emd_19276_additional_1.map.gz emd_19276_additional_1.map.gz emd_19276_half_map_1.map.gz emd_19276_half_map_1.map.gz emd_19276_half_map_2.map.gz emd_19276_half_map_2.map.gz | 15 MB 28.3 MB 28.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19276 http://ftp.pdbj.org/pub/emdb/structures/EMD-19276 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19276 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19276 | HTTPS FTP |
-Validation report
| Summary document |  emd_19276_validation.pdf.gz emd_19276_validation.pdf.gz | 853 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_19276_full_validation.pdf.gz emd_19276_full_validation.pdf.gz | 852.6 KB | Display | |
| Data in XML |  emd_19276_validation.xml.gz emd_19276_validation.xml.gz | 14.1 KB | Display | |
| Data in CIF |  emd_19276_validation.cif.gz emd_19276_validation.cif.gz | 18 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19276 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19276 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19276 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19276 | HTTPS FTP |
-Related structure data
| Related structure data |  8bquC  8rkeC  8rkfC  8rkgC  8rkhC  8rkiC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19276.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19276.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
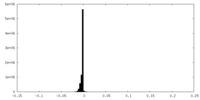
| Annotation | Sharpened cryo-EM map of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) (C1) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.9459 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unprocessed cryo-EM map of tetrameric collagenase-cleaved Xenopus ZP2-N2N3...
| File | emd_19276_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
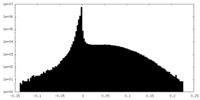
| Annotation | Unprocessed cryo-EM map of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) (C1) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM half map 1 of tetrameric collagenase-cleaved Xenopus...
| File | emd_19276_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
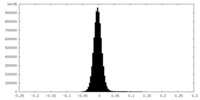
| Annotation | Cryo-EM half map 1 of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) (C1) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM half map 2 of tetrameric collagenase-cleaved Xenopus...
| File | emd_19276_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
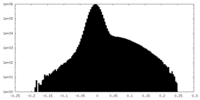
| Annotation | Cryo-EM half map 2 of tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) (C1) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3)
| Entire | Name: Tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) |
|---|---|
| Components |
|
-Supramolecule #1: Tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3)
| Supramolecule | Name: Tetrameric collagenase-cleaved Xenopus ZP2-N2N3 (cleaved xZP2-N2N3) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: |
-Macromolecule #1: Zona pellucida sperm-binding protein 2 (ZP2) / gp69/64
| Macromolecule | Name: Zona pellucida sperm-binding protein 2 (ZP2) / gp69/64 type: protein_or_peptide / ID: 1 Details: The protein consists of two polypeptide fragments, generated by collagenase cleavage between P160 and V161, that remain covalently linked via a disulfide bond between C139 and C244 Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DEPGSSVVTC TKDSMTVRIP RTLSGFDDEI PVAAPSFWDL EVKFTGQTSL LGMSEARQRG YQFSSDPYYL TVQASYSAFG LNVFNLENQR LYVADLRLVS QFGSPRISID TPMICARDSP SCNSTHATVL IPFFGGVLTG INVNSVNIQL SSYSLQQHGI TLDSRNGYRL ...String: DEPGSSVVTC TKDSMTVRIP RTLSGFDDEI PVAAPSFWDL EVKFTGQTSL LGMSEARQRG YQFSSDPYYL TVQASYSAFG LNVFNLENQR LYVADLRLVS QFGSPRISID TPMICARDSP SCNSTHATVL IPFFGGVLTG INVNSVNIQL SSYSLQQHGI TLDSRNGYRL YIKRSTLKGD RNDVLVLTFI YYGKTVPMLI SLVCSGGSNL EHHHHHHHH UniProtKB: Vitelline envelope 69/64 kDa glycoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL |
|---|---|
| Buffer | pH: 7.8 |
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR / Details: 20 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Incubate 30 seconds, blot for 3 seconds, force 0 before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 1347 / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 15000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.4000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)