[English] 日本語
 Yorodumi
Yorodumi- EMDB-19199: Structure of the rabbit 80S ribosome stalled on a 2-TMD rhodopsin... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the rabbit 80S ribosome stalled on a 2-TMD rhodopsin intermediate in complex with Sec61, bound TRAP-alpha TMD | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome / Membrane protein / Translocon / Transport | |||||||||
| Biological species |  | |||||||||
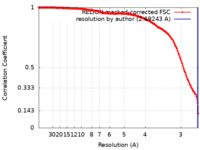
| Method | single particle reconstruction / cryo EM / Resolution: 2.69243 Å | |||||||||
 Authors Authors | Lewis AJO / Hegde RS | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Structural analysis of the dynamic ribosome-translocon complex. Authors: Aaron J O Lewis / Frank Zhong / Robert J Keenan / Ramanujan S Hegde /   Abstract: The protein translocon at the endoplasmic reticulum comprises the Sec61 translocation channel and numerous accessory factors that collectively facilitate the biogenesis of secretory and membrane ...The protein translocon at the endoplasmic reticulum comprises the Sec61 translocation channel and numerous accessory factors that collectively facilitate the biogenesis of secretory and membrane proteins. Here, we leveraged recent advances in cryo-electron microscopy (cryo-EM) and structure prediction to derive insights into several novel configurations of the ribosome-translocon complex. We show how a transmembrane domain (TMD) in a looped configuration passes through the Sec61 lateral gate during membrane insertion; how a nascent chain can bind and constrain the conformation of ribosomal protein uL22; and how the translocon-associated protein (TRAP) complex can adjust its position during different stages of protein biogenesis. Most unexpectedly, we find that a large proportion of translocon complexes contains RAMP4 intercalated into Sec61's lateral gate, widening Sec61's central pore and contributing to its hydrophilic interior. These structures lead to mechanistic hypotheses for translocon function and highlight a remarkably plastic machinery whose conformations and composition adjust dynamically to its diverse range of substrates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19199.map.gz emd_19199.map.gz | 263.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19199-v30.xml emd-19199-v30.xml emd-19199.xml emd-19199.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19199_fsc.xml emd_19199_fsc.xml | 14.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_19199.png emd_19199.png | 146.2 KB | ||
| Filedesc metadata |  emd-19199.cif.gz emd-19199.cif.gz | 4.3 KB | ||
| Others |  emd_19199_half_map_1.map.gz emd_19199_half_map_1.map.gz emd_19199_half_map_2.map.gz emd_19199_half_map_2.map.gz | 224.8 MB 224.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19199 http://ftp.pdbj.org/pub/emdb/structures/EMD-19199 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19199 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19199 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_19199.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19199.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3398 Å | ||||||||||||||||||||||||||||||||||||
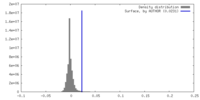
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_19199_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
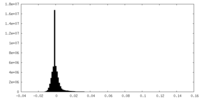
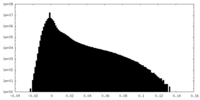
| Density Histograms |
-Half map: #2
| File | emd_19199_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
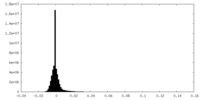
| Density Histograms |
- Sample components
Sample components
-Entire : 80S ribosome translating a stalled, two-TMD nascent chain (derive...
| Entire | Name: 80S ribosome translating a stalled, two-TMD nascent chain (derived from rhodopsin), and bound to Sec61, the TRAP complex, and RAMP4 |
|---|---|
| Components |
|
-Supramolecule #1: 80S ribosome translating a stalled, two-TMD nascent chain (derive...
| Supramolecule | Name: 80S ribosome translating a stalled, two-TMD nascent chain (derived from rhodopsin), and bound to Sec61, the TRAP complex, and RAMP4 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#56 / Details: Sample prepared by in vitro translation |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 17540 / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.9000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)