+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CgsiGP3 sample in nanodisc | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | cgs / cyclic glucan / ANTIBIOTIC | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcellobiose phosphorylase / cellobiose phosphorylase activity / carbohydrate binding / carbohydrate metabolic process / membrane Similarity search - Function | |||||||||||||||
| Biological species |  Agrobacterium tumefaciens (bacteria) Agrobacterium tumefaciens (bacteria) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.35 Å | |||||||||||||||
 Authors Authors | Sedzicki J / Ni D / Lehmann F / Stahlberg H / Dehio C | |||||||||||||||
| Funding support |  Switzerland, 4 items Switzerland, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure-function analysis of the cyclic β-1,2-glucan synthase from Agrobacterium tumefaciens. Authors: Jaroslaw Sedzicki / Dongchun Ni / Frank Lehmann / Henning Stahlberg / Christoph Dehio /  Abstract: The synthesis of complex sugars is a key aspect of microbial biology. Cyclic β-1,2-glucan (CβG) is a circular polysaccharide critical for host interactions of many bacteria, including major ...The synthesis of complex sugars is a key aspect of microbial biology. Cyclic β-1,2-glucan (CβG) is a circular polysaccharide critical for host interactions of many bacteria, including major pathogens of humans (Brucella) and plants (Agrobacterium). CβG is produced by the cyclic glucan synthase (Cgs), a multi-domain membrane protein. So far, its structure as well as the mechanism underlining the synthesis have not been clarified. Here we use cryo-electron microscopy (cryo-EM) and functional approaches to study Cgs from A. tumefaciens. We determine the structure of this complex protein machinery and clarify key aspects of CβG synthesis, revealing a distinct mechanism that uses a tyrosine-linked oligosaccharide intermediate in cycles of polymerization and processing of the glucan chain. Our research opens possibilities for combating pathogens that rely on polysaccharide virulence factors and may lead to synthetic biology approaches for producing complex cyclic sugars. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_19119.map.gz emd_19119.map.gz | 118.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-19119-v30.xml emd-19119-v30.xml emd-19119.xml emd-19119.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_19119_fsc.xml emd_19119_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_19119.png emd_19119.png | 98.6 KB | ||
| Filedesc metadata |  emd-19119.cif.gz emd-19119.cif.gz | 7.4 KB | ||
| Others |  emd_19119_half_map_1.map.gz emd_19119_half_map_1.map.gz emd_19119_half_map_2.map.gz emd_19119_half_map_2.map.gz | 116.2 MB 116.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19119 http://ftp.pdbj.org/pub/emdb/structures/EMD-19119 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19119 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19119 | HTTPS FTP |
-Related structure data
| Related structure data |  8rfgMC  8rf0C  8rf9C  8rfeC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_19119.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_19119.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2812 Å | ||||||||||||||||||||||||||||||||||||
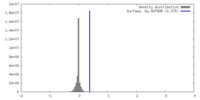
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_19119_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
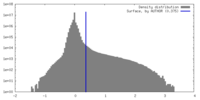
| Density Histograms |
-Half map: #2
| File | emd_19119_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
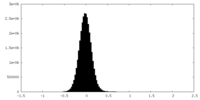
| Density Histograms |
- Sample components
Sample components
-Entire : cyclic b-1,2-glucan synthase from Agrobacterium tumefaciens
| Entire | Name: cyclic b-1,2-glucan synthase from Agrobacterium tumefaciens |
|---|---|
| Components |
|
-Supramolecule #1: cyclic b-1,2-glucan synthase from Agrobacterium tumefaciens
| Supramolecule | Name: cyclic b-1,2-glucan synthase from Agrobacterium tumefaciens type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Agrobacterium tumefaciens (bacteria) Agrobacterium tumefaciens (bacteria) |
-Macromolecule #1: Cyclic beta 1-2 glucan synthetase
| Macromolecule | Name: Cyclic beta 1-2 glucan synthetase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: cellobiose phosphorylase |
|---|---|
| Source (natural) | Organism:  Agrobacterium tumefaciens (bacteria) Agrobacterium tumefaciens (bacteria) |
| Molecular weight | Theoretical: 313.358656 KDa |
| Sequence | String: DHNDSIRASY MTIEELHDAG AALSRDGADS LPGFMEFDFF ERHRENEKEI LRVYRTTAVD AENGATITPA AEWLLDNHYV IEEAIQEVR RDFPRKFYRQ LPTMTVGGVT IPRVMALGWL YVAHTHSTVS RENMTALVDG YQTSQTLQIG ELWALPSIIR F VLIENLRR ...String: DHNDSIRASY MTIEELHDAG AALSRDGADS LPGFMEFDFF ERHRENEKEI LRVYRTTAVD AENGATITPA AEWLLDNHYV IEEAIQEVR RDFPRKFYRQ LPTMTVGGVT IPRVMALGWL YVAHTHSTVS RENMTALVDG YQTSQTLQIG ELWALPSIIR F VLIENLRR ISIRVERSRR MRQKANEVVD EIIRLNDAEA SAALLKQVDS LVDDPTFATQ FLYRLRNGSQ TSGFAVAWLE ER LHAAGTD AENVMMSEHN RLASGNVTMG NIVKSLREID DTEWSVWFEE VSHIDKVLRE ETDYEILDFG SRNTYRNTIE LLA RRSPRT EVEVARAAVE MARSDLPAGA DENHRVNVGS VLVGQRRFEL EKALGYRPLA SQHIVRSMRK FNWLAIAAPV LLIT AVAML AVGWFLAKAG MPWYVVTAFL LMFALPASEG ATGLFNTLVT FFVKPFRLVG YEFKNGIPED ARTLVAVPCM LTSRD SVDE MMRNIEVHYL ANPHGEIYFS LVSDWRDAPY EQSDEDLEIL DYAKRELAAL NSRYAFDGKT RFYLLHRRRI YNSAEE CWM GWERKRGKLH ELNMLLRGDK DTTFLGGSNI VPADVKYVMT LDADTRLMRD AVTKLVGKMH HPINRPKIDP VSGRVVE GY GLLQPRVTPS LTTGKDASVF QRVFSINRGI DPYVFTVSDV YQDLTSEGTF TGKGLYDVDA FEAALKGRIE ENSILSHD L LEGSFARCAL VTDVELVEDF PTRYEVEVSR QHRWARGDWQ LLPFIIDRAR GVTAIGRWKM VDNLRRSLTP IAWFFASIL GWYFMDPLGA LIWQILLIFS LFVAPTLSLL SGLVPRSTDI VPQAHFFTIW SEIRATNAQV ALRIVFIADA ACMMTDAIVR SLYRLLVSH KLMLEWRTAA SMQSSAQGSI VDYYRQMWHA PVVAMLGLLF AALPGDNAFL IGIPFTLLWV LSPAVAWYVS Q SAETEDRL FVSEHVSFEL RKIARRTWRY YEAFVTPQEN HLPPDNFQET PEPIVASRTS PTNIGVYLLS VISARQFGWI SF ADTLERI ENTIQTVEKM EKHRGHLYNW YHTDTLQTLG PRYVSAVDSG NLAGHLIAVS SACRDWAEAP SAHLQGNLDG IGD VAGILR ETLKALPDNR KTLRPLHRRL EERIIGFSNA LASVKREHEF ASIRVINLAV LARDIQKLAT NVDHEVKSAQ SAEV TRWAQ LLVESCEAHI SDSAIDLTNM EPLRQRLASL RDRSRNLAFS MDFTFLYRKD RRLLSIGYRV ESKELDEACY DLLAS ECRL TSLFAIAKGD LPTEHWYRLG RQVVPIGAQG ALVSWSGSMF EYLMPPLVMQ ERQGGILNQT NNLIVKEQMN HGRRLG TPW GISEAAFNAR DHNMNYQYTN FGVPTLGLKR GLGQNAVIAP YASILASQYD PDGALENLDK LRKLGALGQY GFHDAVD FT PTRVPDGKVC AVVYNYYAHH HGMSIAAVAN VAFDGVLREL FHSDPVIEAA ELLLQEKAPR EVPVMSAKYE PETPGKEQ A DLLRAEVRSI ADPAVRDREV VFLSNGHYST MLTSTGAGYS KWNGQAISRW KADPTDDRWG TFIFLRDTTN GQWWSATAE PRVIEGEKTK TIFTDDKAEF HKTIGDLQSV VECIVATEHD AEGRRITLLN VGSEDRYIEV TSYMEPVIAS EDDDNAHPLF SRMFVQTEI GRRGDVIRAW RNRRSQNEPG TVIAHLAADN AGPSRPTEFE TDRAKFIGRG RSLREAAAFD AGATLSSSDG F TLDPILSL RRTVRVPAGK KVSVIFWTIA APSREEVDKA IDRYRHPDAF AHELVHAWTR TQVQMRHVGV TSQQAAAFQH LG RYLTYPD MHLRADSETL KTGLASQRAL WPLAISGDFP IFSLRINDDM DMDIAREALS AHEYLRSRGV IFDLVIVNER AAS YAQDMQ HALDHISETQ RRINPADGGR PHVFSVRRDL MDEETWSALL AASRVVLHVR NGKIVDQINR AVSLFAANRG PDGS SDAAQ ARLPVPAFPV AEPVEDAGDL DFWNGFGGFA KNGQEYVVRL NGGQSTPHPW INVISNENFG FHISAEGAGF SWSRN SRDY QLTPWTNDPV INRPGEAFYV ADVETGKLYT PCAALSRDPE AMFETRHGLG YSILTGVADT LEVELTQTVD REKPVK FSQ VIVRNKGSKS RRLKVYAYVE WVLGNNGQKS APFILSRHDA GSNAIFASNP YSIDYSARTS FLTLDSEASG FTTSRRE FI GRFGSAQAPQ GIVAGAALSG TTEVDGDPCA ALMQEIHLKP GEERHMTFIL GDADNAEEAE ALVKDVRQAD FLSVLEES K KFWTGFTGQL QVSTPDAGFN HMVNNWLPYQ ALACRILART AFYQSSGAFG FRDQLQDTLA FLLYQPDLAR TQILRAAGR QFPEGDVQHW WLPLTGAGVR TTISDDVVWL AYAINQYVSA TGDAAILDES IPFLKGPALM PGQHDAFFQP ETSERSATLY EHAALALDL AIHRTGENGL PLILGGDWND GMNRVGVGGK GTSVWLGWFL AGALRDFIEI AEKRGDTDRV GKWASHREKL R HVLETAGW DGSYYRRGYF DDGTPLGSAS SEECQIDSLG QSWSVLSGEG EEGRSRQAMD AVMEHLVDEK TGIIRLFTPP FS RASHDPG YIKGYPPGVR ENGGQYTHAA TWVVLALAKQ GRAQEAWNCF KLLNPVNHAL DAASSETYRV EPYVVTADVY GEG AYAGRG GWSWYTGSAG WLYRAAVEGI LGITRTDGKL HVSPSLPEDW SGFSIRITLD GKARDIAVSR KAGTADVSVS VDG UniProtKB: Cyclic beta 1-2 glucan synthetase |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 2 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)