[English] 日本語
 Yorodumi
Yorodumi- EMDB-18904: Structure of the Co(II) triggered TRAP (S33HK35H) protein cage (l... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Co(II) triggered TRAP (S33HK35H) protein cage (levo form) | |||||||||
 Map data Map data | half map A | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRAP protein / protein cage / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | Transcription attenuation protein MtrB / Tryptophan RNA-binding attenuator protein domain / Tryptophan RNA-binding attenuator protein / Tryptophan RNA-binding attenuator protein-like domain superfamily / DNA-templated transcription termination / regulation of DNA-templated transcription / RNA binding / identical protein binding / Transcription attenuation protein MtrB Function and homology information Function and homology information | |||||||||
| Biological species |   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) | |||||||||
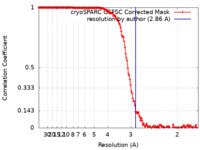
| Method | single particle reconstruction / cryo EM / Resolution: 2.86 Å | |||||||||
 Authors Authors | Biela AP / Heddle JG | |||||||||
| Funding support |  Poland, 1 items Poland, 1 items
| |||||||||
 Citation Citation |  Journal: Macromol Rapid Commun / Year: 2025 Journal: Macromol Rapid Commun / Year: 2025Title: Designed, Programmable Protein Cages Utilizing Diverse Metal Coordination Geometries Show Reversible, pH-Dependent Assembly. Authors: Norbert Osiński / Karolina Majsterkiewicz / Zuzanna Pakosz-Stępień / Yusuke Azuma / Artur P Biela / Szymon Gaweł / Jonathan G Heddle /   Abstract: The rational design and production of a novel series of engineered protein cages are presented, which have emerged as versatile and adaptable platforms with significant applications in biomedicine. ...The rational design and production of a novel series of engineered protein cages are presented, which have emerged as versatile and adaptable platforms with significant applications in biomedicine. These protein cages are assembled from multiple protein subunits, and precise control over their interactions is crucial for regulating assembly and disassembly, such as the on-demand release of encapsulated therapeutic agents. This approach employs a homo-undecameric, ring-shaped protein scaffold with strategically positioned metal binding sites. These engineered proteins can self-assemble into highly stable cages in the presence of cobalt or zinc ions. Furthermore, the cages can be disassembled on demand by employing external triggers such as chelating agents and changes in pH. Interestingly, for certain triggers, the disassembly process is reversible, allowing the cages to reassemble upon reversal or outcompeting of triggering conditions/agents. This work offers a promising platform for the development of advanced drug delivery systems and other biomedical applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18904.map.gz emd_18904.map.gz | 327.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18904-v30.xml emd-18904-v30.xml emd-18904.xml emd-18904.xml | 17 KB 17 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18904_fsc.xml emd_18904_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_18904.png emd_18904.png | 114.4 KB | ||
| Filedesc metadata |  emd-18904.cif.gz emd-18904.cif.gz | 6 KB | ||
| Others |  emd_18904_half_map_1.map.gz emd_18904_half_map_1.map.gz emd_18904_half_map_2.map.gz emd_18904_half_map_2.map.gz | 317.9 MB 317.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18904 http://ftp.pdbj.org/pub/emdb/structures/EMD-18904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18904 | HTTPS FTP |
-Related structure data
| Related structure data |  8r59MC  8r5aC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18904.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18904.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: main map
| File | emd_18904_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_18904_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TRAP(S33HK35C) protein cage
| Entire | Name: TRAP(S33HK35C) protein cage |
|---|---|
| Components |
|
-Supramolecule #1: TRAP(S33HK35C) protein cage
| Supramolecule | Name: TRAP(S33HK35C) protein cage / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) |
| Molecular weight | Theoretical: 2.2 MDa |
-Macromolecule #1: Transcription attenuation protein MtrB
| Macromolecule | Name: Transcription attenuation protein MtrB / type: protein_or_peptide / ID: 1 / Number of copies: 264 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Geobacillus stearothermophilus (bacteria) Geobacillus stearothermophilus (bacteria) |
| Molecular weight | Theoretical: 8.317413 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MYTNSDFVVI KALEDGVNVI GLTRGADTRF HHHEHLDKGE VLIAQFTEHT SAIKVRGKAY IQTRHGVIES EGKK UniProtKB: Transcription attenuation protein MtrB |
-Macromolecule #2: COBALT (II) ION
| Macromolecule | Name: COBALT (II) ION / type: ligand / ID: 2 / Number of copies: 120 / Formula: CO |
|---|---|
| Molecular weight | Theoretical: 58.933 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.9 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 70 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)