+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo EM structure of a stable LGL/aPKC Iota/Par-6 complex | ||||||||||||
 マップデータ マップデータ | Unfiltered map | ||||||||||||
 試料 試料 |
| ||||||||||||
 キーワード キーワード | Kinase / Polarity / Kinase substrate complex. / CYTOSOLIC PROTEIN | ||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報Golgi cis cisterna / regulation of cellular localization / establishment of spindle orientation / diacylglycerol-dependent, calcium-independent serine/threonine kinase activity / Golgi vesicle budding / PAR polarity complex / Tight junction interactions / regulation of establishment or maintenance of cell polarity / cell-cell junction maintenance / protein kinase C ...Golgi cis cisterna / regulation of cellular localization / establishment of spindle orientation / diacylglycerol-dependent, calcium-independent serine/threonine kinase activity / Golgi vesicle budding / PAR polarity complex / Tight junction interactions / regulation of establishment or maintenance of cell polarity / cell-cell junction maintenance / protein kinase C / establishment of apical/basal cell polarity / diacylglycerol-dependent serine/threonine kinase activity / myosin II binding / regulation of Notch signaling pathway / negative regulation of glial cell apoptotic process / eye photoreceptor cell development / positive regulation of protein localization to centrosome / Schmidt-Lanterman incisure / GTP-dependent protein binding / establishment or maintenance of epithelial cell apical/basal polarity / Golgi to plasma membrane transport / cellular response to chemical stress / membrane organization / cell-cell junction organization / centrosome cycle / cortical actin cytoskeleton organization / protein targeting to membrane / tight junction / RHOV GTPase cycle / cortical actin cytoskeleton / positive regulation of Notch signaling pathway / establishment of cell polarity / establishment or maintenance of cell polarity / cell leading edge / exocytosis / RHOU GTPase cycle / brush border / CDC42 GTPase cycle / positive regulation of glial cell proliferation / positive regulation of endothelial cell apoptotic process / viral process / bicellular tight junction / regulation of postsynaptic membrane neurotransmitter receptor levels / intercellular bridge / vesicle-mediated transport / ruffle / RAC1 GTPase cycle / cytoskeleton organization / p75NTR recruits signalling complexes / secretion / response to interleukin-1 / GTPase activator activity / axonogenesis / actin filament organization / trans-Golgi network membrane / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) / positive regulation of protein secretion / positive regulation of D-glucose import / protein localization to plasma membrane / Asymmetric localization of PCP proteins / positive regulation of protein localization to plasma membrane / adherens junction / positive regulation of NF-kappaB transcription factor activity / positive regulation of neuron projection development / phospholipid binding / small GTPase binding / Pre-NOTCH Transcription and Translation / centriolar satellite / Schaffer collateral - CA1 synapse / cellular response to insulin stimulus / KEAP1-NFE2L2 pathway / cell migration / microtubule cytoskeleton / protein-containing complex assembly / early endosome membrane / cell cortex / negative regulation of neuron apoptotic process / cytoskeleton / protein phosphorylation / protein kinase activity / endosome / intracellular signal transduction / cilium / apical plasma membrane / Golgi membrane / axon / cell division / protein serine kinase activity / intracellular membrane-bounded organelle / protein serine/threonine kinase activity / centrosome / protein kinase binding / negative regulation of apoptotic process / glutamatergic synapse / structural molecule activity / extracellular exosome / zinc ion binding / nucleoplasm / ATP binding / nucleus 類似検索 - 分子機能 | ||||||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | ||||||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.68 Å | ||||||||||||
 データ登録者 データ登録者 | Earl CP / Briggs DC / McDonald NQ | ||||||||||||
| 資金援助 |  英国, 3件 英国, 3件
| ||||||||||||
 引用 引用 |  ジャーナル: Nat Struct Mol Biol / 年: 2025 ジャーナル: Nat Struct Mol Biol / 年: 2025タイトル: Capture, mutual inhibition and release mechanism for aPKC-Par6 and its multisite polarity substrate Lgl. 著者: Christopher P Earl / Mathias Cobbaut / André Barros-Carvalho / Marina E Ivanova / David C Briggs / Eurico Morais-de-Sá / Peter J Parker / Neil Q McDonald /   要旨: The mutually antagonistic relationship of atypical protein kinase C (aPKC) and partitioning-defective protein 6 (Par6) with the substrate lethal (2) giant larvae (Lgl) is essential for regulating ...The mutually antagonistic relationship of atypical protein kinase C (aPKC) and partitioning-defective protein 6 (Par6) with the substrate lethal (2) giant larvae (Lgl) is essential for regulating polarity across many cell types. Although aPKC-Par6 phosphorylates Lgl at three serine sites to exclude it from the apical domain, aPKC-Par6 and Lgl paradoxically form a stable kinase-substrate complex, with conflicting roles proposed for Par6. We report the structure of human aPKCι-Par6α bound to full-length Llgl1, captured through an aPKCι docking site and a Par6 contact. This complex traps a phospho-S663 Llgl1 intermediate bridging between aPKC and Par6, impeding phosphorylation progression. Thus, aPKCι is effectively inhibited by Llgl1 while Llgl1 is captured by aPKCι-Par6. Mutational disruption of the Lgl-aPKC interaction impedes complex assembly and Lgl phosphorylation, whereas disrupting the Lgl-Par6 contact promotes complex dissociation and Lgl phosphorylation. We demonstrate a Par6-regulated substrate capture-and-release model requiring binding by active Cdc42 and the apical partner Crumbs to drive complex disassembly. Our results suggest a mechanism for mutual regulation and spatial control of aPKC-Par6 and Lgl activities. | ||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_18877.map.gz emd_18877.map.gz | 2.9 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-18877-v30.xml emd-18877-v30.xml emd-18877.xml emd-18877.xml | 27.1 KB 27.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_18877_fsc.xml emd_18877_fsc.xml | 10 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_18877.png emd_18877.png | 62.7 KB | ||
| Filedesc metadata |  emd-18877.cif.gz emd-18877.cif.gz | 8.6 KB | ||
| その他 |  emd_18877_additional_1.map.gz emd_18877_additional_1.map.gz emd_18877_half_map_1.map.gz emd_18877_half_map_1.map.gz emd_18877_half_map_2.map.gz emd_18877_half_map_2.map.gz | 5.9 MB 77.6 MB 77.6 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18877 http://ftp.pdbj.org/pub/emdb/structures/EMD-18877 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18877 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18877 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_18877_validation.pdf.gz emd_18877_validation.pdf.gz | 1.1 MB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_18877_full_validation.pdf.gz emd_18877_full_validation.pdf.gz | 1.1 MB | 表示 | |
| XML形式データ |  emd_18877_validation.xml.gz emd_18877_validation.xml.gz | 17.2 KB | 表示 | |
| CIF形式データ |  emd_18877_validation.cif.gz emd_18877_validation.cif.gz | 22.6 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18877 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18877 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18877 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18877 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8r3yMC  8r3xC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_18877.map.gz / 形式: CCP4 / 大きさ: 83.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_18877.map.gz / 形式: CCP4 / 大きさ: 83.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Unfiltered map | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-追加マップ: Phenix Sharpened Map used for Model building
| ファイル | emd_18877_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Phenix Sharpened Map used for Model building | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Half Map A
| ファイル | emd_18877_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half Map A | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Half Map B
| ファイル | emd_18877_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Half Map B | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : aPKCiota-Par6-Llgl1 complex
| 全体 | 名称: aPKCiota-Par6-Llgl1 complex |
|---|---|
| 要素 |
|
-超分子 #1: aPKCiota-Par6-Llgl1 complex
| 超分子 | 名称: aPKCiota-Par6-Llgl1 complex / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#3 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 220 KDa |
-分子 #1: Protein kinase C iota type
| 分子 | 名称: Protein kinase C iota type / タイプ: protein_or_peptide / ID: 1 / コピー数: 1 / 光学異性体: LEVO / EC番号: protein kinase C |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 39.146004 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: SLGLQDFDLL RVIGRGSYAK VLLVRLKKTD RIYAMKVVKK ELVNDDEDID WVQTEKHVFE QASNHPFLVG LHSCFQTESR LFFVIEYVN GGDLMFHMQR QRKLPEEHAR FYSAEISLAL NYLHERGIIY RDLKLDNVLL DSEGHIKLTD YGMCKEGLRP G DTTS(TPO) ...文字列: SLGLQDFDLL RVIGRGSYAK VLLVRLKKTD RIYAMKVVKK ELVNDDEDID WVQTEKHVFE QASNHPFLVG LHSCFQTESR LFFVIEYVN GGDLMFHMQR QRKLPEEHAR FYSAEISLAL NYLHERGIIY RDLKLDNVLL DSEGHIKLTD YGMCKEGLRP G DTTS(TPO)FCG TPNYIAPEIL RGEDYGFSVD WWALGVLMFE MMAGRSPFDI VGSSDNPDQN TEDYLFQVIL EKQIRIPR S LSVKAASVLK SFLNKDPKER LGCHPQTGFA DIQGHPFFRN VDWDMMEQKQ VVPPFKPNIS GEFGLDNFDS QFTNEPVQL (TPO)PDDDDIVRK IDQSEFEGFE YI UniProtKB: Protein kinase C iota type |
-分子 #2: Lethal(2) giant larvae protein homolog 1
| 分子 | 名称: Lethal(2) giant larvae protein homolog 1 / タイプ: protein_or_peptide / ID: 2 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 101.889805 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: REKLKQELFA FNKTVEHGFP NQPSALAFDP ELRIMAIGTR SGAVKIYGAP GVEFTGLHRD AATVTQMHFL TGQGRLLSLL DDSSLHLWE IVHHNGCAHL EEALSFQLPS RPGFDGASAP LSLTRVTVVL LVAAGDIAAL GTEGSSVFFL DVTTLTLLEG Q TLAPGEVL ...文字列: REKLKQELFA FNKTVEHGFP NQPSALAFDP ELRIMAIGTR SGAVKIYGAP GVEFTGLHRD AATVTQMHFL TGQGRLLSLL DDSSLHLWE IVHHNGCAHL EEALSFQLPS RPGFDGASAP LSLTRVTVVL LVAAGDIAAL GTEGSSVFFL DVTTLTLLEG Q TLAPGEVL RSVPDDYRCG KALGPVESLQ GHLRDPTKIL IGYSRGLLVI WNQASQCVDH IFLGNQQLES LCWGRDSSTV VS SHSDGSY AVWSVDAGSF PTLQPTVATT PYGPFPCKAI NKILWRNCES GGHFIIFSGG MPRASYGDRH CVSVLRAETL VTL DFTSRI IDFFTVHSTR PEDEFDDPQA LAVLLEEELV VLDLQTPGWP AVPAPYLAPL HSSAITCSAH VASVPAKLWA RIVS AGEQQ SPQPVSSALS WPITGGRNLA QEPSQRGLLL TGHEDGTVRF WDASGVALRP LYKLSTAGLF QTDCEHADSL AQAAE DDWP PFRKVGCFDP YSDDPRLGVQ KVALCKYTAQ MVVAGTAGQV LVLELSDVPV EQAVSVAIID LLQDREGFTW KGHERL SPR TGPLPWPAGF QPRVLVQCLP PAAVTAVTLH TEWSLVAFGT SHGFGLFDYQ RKSPVLARCT LHPNDSLAME GPLSRVK SL KKSLRQ(SEP)FRR IRKSRVSGKK RAANASSKLQ EANAQLAEQA CPHDVEMTPV QRRIEPRSAD DSLSGVVRCL YFAD TFLRD GAHHGPTMWA GTNSGSVFAY ALEVPAAAVG GEKRPEQAVE AVLGKEVQLM HRAPVVAIAV LDGRGRPLPE PYEAS RDLA QAPDMQGGHA VLIASEEQFK VFTLPKVSAK TKFKLTAHEG CRVRKVALAT FASVACEDYA ETCLACLTNL GDVHVF SVP GLRPQVHYSC IRKEDISGIA SCVFTRHGQG FYLISPSEFE RFSLSARNIT EPLCSLDI UniProtKB: Lethal(2) giant larvae protein homolog 1 |
-分子 #3: Partitioning defective 6 homolog alpha
| 分子 | 名称: Partitioning defective 6 homolog alpha / タイプ: protein_or_peptide / ID: 3 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 10.856517 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: THRRVRLHKH GSDRPLGFYI RDGMSVRVAP QGLERVPGIF ISRLVRGGLA ESTGLLAVSD EILEVNGIEV AGKTLDQVTD MMVANSHNL IVTVKPANQR UniProtKB: Partitioning defective 6 homolog alpha |
-分子 #4: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| 分子 | 名称: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / タイプ: ligand / ID: 4 / コピー数: 1 / 式: ANP |
|---|---|
| 分子量 | 理論値: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.4 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 7.5 構成要素:
| ||||||||
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: COPPER / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 45 sec. / 詳細: 45mA | ||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 298 K / 装置: FEI VITROBOT MARK IV 詳細: 4 ul of aPKCiota-Par6-Llgl1 complex at a concentration of 0.4 mg/ml was applied to R1.2/1.3 Quantifoil 300 mesh copper grids which had been glow-discharged for 45 s at 45 mA . Grids were ...詳細: 4 ul of aPKCiota-Par6-Llgl1 complex at a concentration of 0.4 mg/ml was applied to R1.2/1.3 Quantifoil 300 mesh copper grids which had been glow-discharged for 45 s at 45 mA . Grids were blotted for 2.5 s at 100% humidity using an FEI Vitrobot MK IV.. | ||||||||
| 詳細 | Sample was double affinity purified and gel filtered. Sample was monodisperse. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 特殊光学系 | エネルギーフィルター - 名称: GIF Quantum ER |
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / 実像数: 4002 / 平均露光時間: 8.0 sec. / 平均電子線量: 48.1 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 3.0 µm / 最小 デフォーカス(公称値): 1.2 µm |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 初期モデル |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 詳細 | Initial fitting was done in Chimera. Interactive Model building was done in COOT & Isolde Refinement was done using Phenix. | ||||||||||||
| 精密化 | 空間: REAL / プロトコル: FLEXIBLE FIT | ||||||||||||
| 得られたモデル |  PDB-8r3y: |
 ムービー
ムービー コントローラー
コントローラー



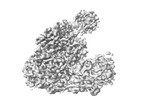















 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)