+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of C. thermophilum RNA exosome core | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nuclease / RNA degradation / RNA metabolism / RNA binding / RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationCUT catabolic process / U1 snRNA 3'-end processing / U5 snRNA 3'-end processing / TRAMP-dependent tRNA surveillance pathway / cytoplasmic exosome (RNase complex) / U4 snRNA 3'-end processing / nuclear polyadenylation-dependent rRNA catabolic process / poly(A)-dependent snoRNA 3'-end processing / nuclear exosome (RNase complex) / exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) ...CUT catabolic process / U1 snRNA 3'-end processing / U5 snRNA 3'-end processing / TRAMP-dependent tRNA surveillance pathway / cytoplasmic exosome (RNase complex) / U4 snRNA 3'-end processing / nuclear polyadenylation-dependent rRNA catabolic process / poly(A)-dependent snoRNA 3'-end processing / nuclear exosome (RNase complex) / exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / nuclear mRNA surveillance / rRNA catabolic process / mRNA 3'-UTR AU-rich region binding / RNA processing / rRNA processing / nucleolus / RNA binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Thermochaetoides thermophila DSM 1495 (fungus) Thermochaetoides thermophila DSM 1495 (fungus) | |||||||||
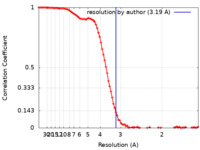
| Method | single particle reconstruction / cryo EM / Resolution: 3.19 Å | |||||||||
 Authors Authors | Lazzaretti D / Liebau J / Pilsl M / Sprangers R | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: 4D structural biology-quantitative dynamics in the eukaryotic RNA exosome complex. Authors: Jobst Liebau / Daniela Lazzaretti / Torben Fürtges / Anna Bichler / Michael Pilsl / Till Rudack / Remco Sprangers /  Abstract: Molecular machines play pivotal roles in all biological processes. Most structural methods, however, are unable to directly probe molecular motions. Here, we demonstrate that dedicated NMR ...Molecular machines play pivotal roles in all biological processes. Most structural methods, however, are unable to directly probe molecular motions. Here, we demonstrate that dedicated NMR experiments can provide quantitative insights into functionally important dynamic regions in very large asymmetric protein complexes. We establish this for the 410 kDa eukaryotic RNA exosome complex that contains ten distinct protein chains. Methyl-group and fluorine NMR experiments reveal site-specific interactions among subunits and with an RNA substrate. Furthermore, we extract quantitative insights into conformational changes within the complex in response to substrate and subunit binding for regions that are invisible in static cryo-EM and crystal structures. In particular, we identify a flexible plug region that can block an aberrant route for RNA towards the active site. Based on molecular dynamics simulations and NMR data, we provide a model that shows how the flexible plug is structured in the open and closed conformations. Our work thus demonstrates that a combination of state-of-the-art structural biology methods can provide quantitative insights into large molecular machines that go significantly beyond the well-resolved and static images of biomolecular complexes, thereby adding the time domain to structural biology. #1:  Journal: Biorxiv / Year: 2024 Journal: Biorxiv / Year: 2024Title: Beyond static structures: quantitative dynamics in the eukaryotic RNA exosome complex Authors: Liebau J / Lazzaretti D / Bichler A / Pilsl M / Sprangers R | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18825.map.gz emd_18825.map.gz | 49.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18825-v30.xml emd-18825-v30.xml emd-18825.xml emd-18825.xml | 35.6 KB 35.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18825_fsc.xml emd_18825_fsc.xml | 11.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_18825.png emd_18825.png | 92.5 KB | ||
| Filedesc metadata |  emd-18825.cif.gz emd-18825.cif.gz | 9.3 KB | ||
| Others |  emd_18825_additional_1.map.gz emd_18825_additional_1.map.gz emd_18825_half_map_1.map.gz emd_18825_half_map_1.map.gz emd_18825_half_map_2.map.gz emd_18825_half_map_2.map.gz | 56.6 MB 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18825 http://ftp.pdbj.org/pub/emdb/structures/EMD-18825 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18825 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18825 | HTTPS FTP |
-Related structure data
| Related structure data |  8r1oMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18825.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18825.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.772 Å | ||||||||||||||||||||||||||||||||||||
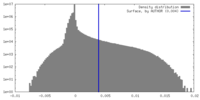
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Post-processed map (deepEMhancer)
| File | emd_18825_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed map (deepEMhancer) | ||||||||||||
| Projections & Slices |
| ||||||||||||
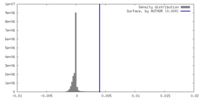
| Density Histograms |
-Half map: #2
| File | emd_18825_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18825_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
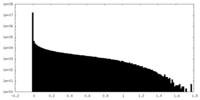
| Density Histograms |
- Sample components
Sample components
+Entire : RNA exosome core (Exo9)
+Supramolecule #1: RNA exosome core (Exo9)
+Macromolecule #1: Rrp45
+Macromolecule #2: Exoribonuclease phosphorolytic domain-containing protein
+Macromolecule #3: Exoribonuclease-like protein
+Macromolecule #4: Exoribonuclease phosphorolytic domain-containing protein
+Macromolecule #5: Exoribonuclease phosphorolytic domain-containing protein
+Macromolecule #6: Exosome complex component MTR3
+Macromolecule #7: Ribosomal RNA-processing protein 40
+Macromolecule #8: Putative exosome complex protein
+Macromolecule #9: Putative exosome 3'->5 protein
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.53 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 20 mM Na-Phosphate buffer pH 7.5, 300 mM NaCl, 1 mM DTT | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 200 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa Details: Grids were glow discharged twice at 15 mA, 0.39 mBar, for 100 seconds each time | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: Sample volume 3 ul Wait time 5 s, Blot time 5 s, Delay time 0 s Force 12. | |||||||||||||||
| Details | Sample frozen directly after S200 size exclusion column |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 200 |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 6579 / Average exposure time: 6.5 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL | |||||||||
| Output model |  PDB-8r1o: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)