+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Apo ReChb , Rec1 improved | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cas / BIOSYNTHETIC PROTEIN / cryo-EM / ancestral sequence reconstruction / biotechnology | |||||||||
| Biological species | synthetic construct (others) | |||||||||
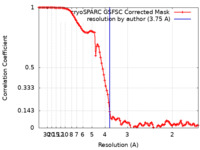
| Method | single particle reconstruction / cryo EM / Resolution: 3.75 Å | |||||||||
 Authors Authors | Lopez-Alonso JP / Tascon I / Ubarretxena-Belandia I | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Biotechnol / Year: 2025 Journal: Nat Biotechnol / Year: 2025Title: A resurrected ancestor of Cas12a expands target access and substrate recognition for nucleic acid editing and detection. Authors: Ylenia Jabalera / Igor Tascón / Sara Samperio / Jorge P López-Alonso / Monika Gonzalez-Lopez / Ana M Aransay / Guillermo Abascal-Palacios / Chase L Beisel / Iban Ubarretxena-Belandia / Raul Perez-Jimenez /   Abstract: The properties of Cas12a nucleases constrict the range of accessible targets and their applications. In this study, we applied ancestral sequence reconstruction (ASR) to a set of Cas12a orthologs ...The properties of Cas12a nucleases constrict the range of accessible targets and their applications. In this study, we applied ancestral sequence reconstruction (ASR) to a set of Cas12a orthologs from hydrobacteria to reconstruct a common ancestor, ReChb, characterized by near-PAMless targeting and the recognition of diverse nucleic acid activators and collateral substrates. ReChb shares 53% sequence identity with the closest Cas12a ortholog but no longer requires a T-rich PAM and can achieve genome editing in human cells at sites inaccessible to the natural FnCas12a or the engineered and PAM-flexible enAsCas12a. Furthermore, ReChb can be triggered not only by double-stranded DNA but also by single-stranded RNA and DNA targets, leading to non-specific collateral cleavage of all three nucleic acid substrates with similar efficiencies. Finally, tertiary and quaternary structures of ReChb obtained by cryogenic electron microscopy reveal the molecular details underlying its expanded biophysical activities. Overall, ReChb expands the application space of Cas12a nucleases and underscores the potential of ASR for enhancing CRISPR technologies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18692.map.gz emd_18692.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18692-v30.xml emd-18692-v30.xml emd-18692.xml emd-18692.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18692_fsc.xml emd_18692_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_18692.png emd_18692.png | 125.1 KB | ||
| Masks |  emd_18692_msk_1.map emd_18692_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18692.cif.gz emd-18692.cif.gz | 6.1 KB | ||
| Others |  emd_18692_half_map_1.map.gz emd_18692_half_map_1.map.gz emd_18692_half_map_2.map.gz emd_18692_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18692 http://ftp.pdbj.org/pub/emdb/structures/EMD-18692 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18692 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18692 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18692.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18692.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8238 Å | ||||||||||||||||||||||||||||||||||||
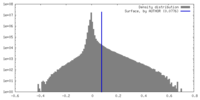
| Density |
| ||||||||||||||||||||||||||||||||||||
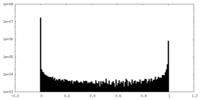
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18692_msk_1.map emd_18692_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
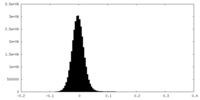
| Density Histograms |
-Half map: #1
| File | emd_18692_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
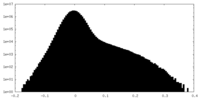
| Density Histograms |
-Half map: #2
| File | emd_18692_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
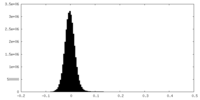
| Density Histograms |
- Sample components
Sample components
-Entire : apo ReChb
| Entire | Name: apo ReChb |
|---|---|
| Components |
|
-Supramolecule #1: apo ReChb
| Supramolecule | Name: apo ReChb / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: ReChb
| Macromolecule | Name: ReChb / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism:  |
| Sequence | String: GPKMFSNFTN QYPLSKTLRF ELKPVGKTLE HIEKKGLLEQ DEKRAEDYKK VKKIIDEYHK DFIEEALNNV KLNGEGLEEY YELYFKKNKD DKDKKKKEFE KIQDNLRKQI VEAFKNHEKY KNLFKKELIK EDLPNWLKNS EDTGEEDKET VEKFKNFTTY FTGFHENRKN ...String: GPKMFSNFTN QYPLSKTLRF ELKPVGKTLE HIEKKGLLEQ DEKRAEDYKK VKKIIDEYHK DFIEEALNNV KLNGEGLEEY YELYFKKNKD DKDKKKKEFE KIQDNLRKQI VEAFKNHEKY KNLFKKELIK EDLPNWLKNS EDTGEEDKET VEKFKNFTTY FTGFHENRKN MYSDEEKSTA IAYRLIHENL PKFLDNMKVF EKIKEKHPEA EQLEKTLKDL GEEEVLGGNN VEDIFSLDYF NHTLTQSGID IYNTIIGGKT EEDGNKKIQG LNEYINLYRQ KNNEKNRKLP KLKPLYKQIL SDRESLSFIP EAFENDEELL EAIEEFYENL NFSNNNEATN VLEKLKELLS NLADYDLNKI YIRNDTSLTD ISQKIFGDWS VIKDALNAHY DQTYPKKKKK KSKEKLEEKR EKWLKKQKYF SIAELQEALD SYCKESDESK EQKENSIADY FKTLAQTKNE TDKKTDLIEN IKSKYQYPND KKLAQDKEFK DVEKIKAFLD SIMNLQHFVK PLHLVKGGSA GAEMEKDEAF YSEFEALYEE LSQVIPLYNK VRNYLTQKPY STEKIKLNFE NSTLLDGWDV NKETDNTSVL LRKDGLYYLG IMNKKHNKVF ENIPESNEND KCYEKMDYKL LPGANKMLPK VFFSNKNIDY FNPSAEILEI YENGTHKKSG DNFNLDDCHK LIDFFKESIN KHEDWKKFGF KFSPTSSYED ISGFYREVEQ QGYKISFKNI SESYIDELVD EGKLYLFQIY NKDFSPYSKG KPNLHTLYWK ALFDEENLKD VVYKLNGEAE VFYRKASINE TIVHKANEPI KNKNPLNPKK QSTFEYDIIK DRRYTVDKFQ FHVPITMNFK AEGNSNINDE VNEFLKGNAP DVNIIGIDRG ERHLLYLTLI DQKGKIVEQD SLNTITNEHN ETDYHALLDD KEKERDKARK SWGTIENIKE LKEGYLSQVV HKIAKLMVEH NAIVVMEDLN FGFKRGRFKV EKQVYQKFEK MLIDKLNYLV DKDKEPNEPG GLLNAYQLTN KFESFQKMGK QSGFLFYVPA WNTSKIDPTT GFVNLFHPRY ENVEKAKEFF NKFDSIRYNS EKDYFEFAFD YNNFTEKAEG TKWTVCTYGE RIKTYRNADK NNQWDSKEVN VTEEFKNLFD EYNIDYKNGN DLKEAILSQD DADFFKSLLH LLRLTLQMRN SITGTEIDYI ISPVANENGE FFDSRKADES LPKDADANGA YHIARKGLWV LEQIKQTDDL KKVNLAISNK EWLEFVQERK N |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 14874 / Average exposure time: 2.1 sec. / Average electron dose: 49.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)