+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Apo ReChb | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cas / BIOSYNTHETIC PROTEIN / cryo-EM / ancestral sequence reconstruction / biotechnology | |||||||||
| Biological species | synthetic construct (others) | |||||||||
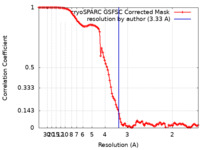
| Method | single particle reconstruction / cryo EM / Resolution: 3.33 Å | |||||||||
 Authors Authors | Lopez-Alonso JP / Ubarretxena-Belandia I / Tascon I | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Biotechnol / Year: 2025 Journal: Nat Biotechnol / Year: 2025Title: A resurrected ancestor of Cas12a expands target access and substrate recognition for nucleic acid editing and detection. Authors: Ylenia Jabalera / Igor Tascón / Sara Samperio / Jorge P López-Alonso / Monika Gonzalez-Lopez / Ana M Aransay / Guillermo Abascal-Palacios / Chase L Beisel / Iban Ubarretxena-Belandia / Raul Perez-Jimenez /   Abstract: The properties of Cas12a nucleases constrict the range of accessible targets and their applications. In this study, we applied ancestral sequence reconstruction (ASR) to a set of Cas12a orthologs ...The properties of Cas12a nucleases constrict the range of accessible targets and their applications. In this study, we applied ancestral sequence reconstruction (ASR) to a set of Cas12a orthologs from hydrobacteria to reconstruct a common ancestor, ReChb, characterized by near-PAMless targeting and the recognition of diverse nucleic acid activators and collateral substrates. ReChb shares 53% sequence identity with the closest Cas12a ortholog but no longer requires a T-rich PAM and can achieve genome editing in human cells at sites inaccessible to the natural FnCas12a or the engineered and PAM-flexible enAsCas12a. Furthermore, ReChb can be triggered not only by double-stranded DNA but also by single-stranded RNA and DNA targets, leading to non-specific collateral cleavage of all three nucleic acid substrates with similar efficiencies. Finally, tertiary and quaternary structures of ReChb obtained by cryogenic electron microscopy reveal the molecular details underlying its expanded biophysical activities. Overall, ReChb expands the application space of Cas12a nucleases and underscores the potential of ASR for enhancing CRISPR technologies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18691.map.gz emd_18691.map.gz | 117.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18691-v30.xml emd-18691-v30.xml emd-18691.xml emd-18691.xml | 24.7 KB 24.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18691_fsc.xml emd_18691_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_18691.png emd_18691.png | 126 KB | ||
| Masks |  emd_18691_msk_1.map emd_18691_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18691.cif.gz emd-18691.cif.gz | 7.6 KB | ||
| Others |  emd_18691_half_map_1.map.gz emd_18691_half_map_1.map.gz emd_18691_half_map_2.map.gz emd_18691_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18691 http://ftp.pdbj.org/pub/emdb/structures/EMD-18691 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18691 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18691 | HTTPS FTP |
-Related structure data
| Related structure data |  8qwdMC  8qweC  8qwfC C: citing same article ( M: atomic model generated by this map |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18691.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18691.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8238 Å | ||||||||||||||||||||||||||||||||||||
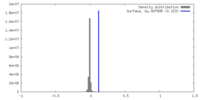
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18691_msk_1.map emd_18691_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
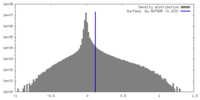
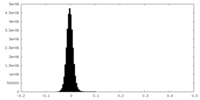
| Density Histograms |
-Half map: #2
| File | emd_18691_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
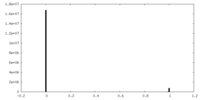
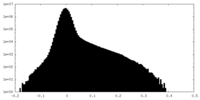
| Density Histograms |
-Half map: #1
| File | emd_18691_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
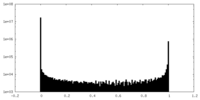
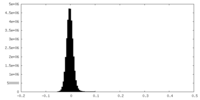
| Density Histograms |
- Sample components
Sample components
-Entire : Apo ReChb
| Entire | Name: Apo ReChb |
|---|---|
| Components |
|
-Supramolecule #1: Apo ReChb
| Supramolecule | Name: Apo ReChb / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: ReChb
| Macromolecule | Name: ReChb / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 148.954109 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPKMFSNFTN QYPLSKTLRF ELKPVGKTLE HIEKKGLLEQ DEKRAEDYKK VKKIIDEYHK DFIEEALNNV KLNGEGLEEY YELYFKKNK DDKDKKKKEF EKIQDNLRKQ IVEAFKNHEK YKNLFKKELI KEDLPNWLKN SEDTGEEDKE TVEKFKNFTT Y FTGFHENR ...String: GPKMFSNFTN QYPLSKTLRF ELKPVGKTLE HIEKKGLLEQ DEKRAEDYKK VKKIIDEYHK DFIEEALNNV KLNGEGLEEY YELYFKKNK DDKDKKKKEF EKIQDNLRKQ IVEAFKNHEK YKNLFKKELI KEDLPNWLKN SEDTGEEDKE TVEKFKNFTT Y FTGFHENR KNMYSDEEKS TAIAYRLIHE NLPKFLDNMK VFEKIKEKHP EAEQLEKTLK DLGEEEVLGG NNVEDIFSLD YF NHTLTQS GIDIYNTIIG GKTEEDGNKK IQGLNEYINL YRQKNNEKNR KLPKLKPLYK QILSDRESLS FIPEAFENDE ELL EAIEEF YENLNFSNNN EATNVLEKLK ELLSNLADYD LNKIYIRNDT SLTDISQKIF GDWSVIKDAL NAHYDQTYPK KKKK KSKEK LEEKREKWLK KQKYFSIAEL QEALDSYCKE SDESKEQKEN SIADYFKTLA QTKNETDKKT DLIENIKSKY QYPND KKLA QDKEFKDVEK IKAFLDSIMN LQHFVKPLHL VKGGSAGAEM EKDEAFYSEF EALYEELSQV IPLYNKVRNY LTQKPY STE KIKLNFENST LLDGWDVNKE TDNTSVLLRK DGLYYLGIMN KKHNKVFENI PESNENDKCY EKMDYKLLPG ANKMLPK VF FSNKNIDYFN PSAEILEIYE NGTHKKSGDN FNLDDCHKLI DFFKESINKH EDWKKFGFKF SPTSSYEDIS GFYREVEQ Q GYKISFKNIS ESYIDELVDE GKLYLFQIYN KDFSPYSKGK PNLHTLYWKA LFDEENLKDV VYKLNGEAEV FYRKASINE TIVHKANEPI KNKNPLNPKK QSTFEYDIIK DRRYTVDKFQ FHVPITMNFK AEGNSNINDE VNEFLKGNAP DVNIIGIDRG ERHLLYLTL IDQKGKIVEQ DSLNTITNEH NETDYHALLD DKEKERDKAR KSWGTIENIK ELKEGYLSQV VHKIAKLMVE H NAIVVMED LNFGFKRGRF KVEKQVYQKF EKMLIDKLNY LVDKDKEPNE PGGLLNAYQL TNKFESFQKM GKQSGFLFYV PA WNTSKID PTTGFVNLFH PRYENVEKAK EFFNKFDSIR YNSEKDYFEF AFDYNNFTEK AEGTKWTVCT YGERIKTYRN ADK NNQWDS KEVNVTEEFK NLFDEYNIDY KNGNDLKEAI LSQDDADFFK SLLHLLRLTL QMRNSITGTE IDYIISPVAN ENGE FFDSR KADESLPKDA DANGAYHIAR KGLWVLEQIK QTDDLKKVNL AISNKEWLEF VQERKN |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 4 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 14874 / Average exposure time: 2.1 sec. / Average electron dose: 49.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: other / Details: ModelAngelo |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-8qwd: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)