[English] 日本語
 Yorodumi
Yorodumi- EMDB-18661: Structure of the Bacteriophage PhiKZ non-virion RNA Polymerase bo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Bacteriophage PhiKZ non-virion RNA Polymerase bound to DNA and RNA | |||||||||
 Map data Map data | PhiKZ nvRNAP DNA/RNA complex Postprocessed Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PhiKZ / nvRNAP / RNA / DNA / transcription / RNA BINDING PROTEIN | |||||||||
| Function / homology | PHIKZ123 / PHIKZ074 / PHIKZ068 / PHIKZ055 Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas phage phiKZ (virus) Pseudomonas phage phiKZ (virus) | |||||||||
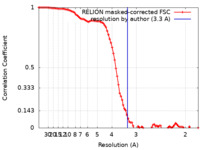
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | de Martin Garrido N / Yakunina M / Aylett CHS | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2024 Journal: J Mol Biol / Year: 2024Title: Structure of the Bacteriophage PhiKZ Non-virion RNA Polymerase Transcribing from its Promoter p119L. Authors: Natàlia de Martín Garrido / Chao-Sheng Chen / Kailash Ramlaul / Christopher H S Aylett / Maria Yakunina /   Abstract: Bacteriophage ΦKZ (PhiKZ) is the founding member of a family of giant bacterial viruses. It has potential as a therapeutic as its host, Pseudomonas aeruginosa, kills tens of thousands of people ...Bacteriophage ΦKZ (PhiKZ) is the founding member of a family of giant bacterial viruses. It has potential as a therapeutic as its host, Pseudomonas aeruginosa, kills tens of thousands of people worldwide each year. ΦKZ infection is independent of the host transcriptional apparatus; the virus forms a "nucleus", producing a proteinaceous barrier around the ΦKZ genome that excludes the host immune systems. It expresses its own non-canonical multi-subunit non-virion RNA polymerase (nvRNAP), which is imported into its "nucleus" to transcribe viral genes. The ΦKZ nvRNAP is formed by four polypeptides representing homologues of the eubacterial β/β' subunits, and a fifth that is likely to have evolved from an ancestral homologue to σ-factor. We have resolved the structure of the ΦKZ nvRNAP initiating transcription from its cognate promoter, p119L, including previously disordered regions. Our results shed light on the similarities and differences between ΦKZ nvRNAP mechanisms of transcription and those of canonical eubacterial RNAPs and the related non-canonical nvRNAP of bacteriophage AR9. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18661.map.gz emd_18661.map.gz | 59.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18661-v30.xml emd-18661-v30.xml emd-18661.xml emd-18661.xml | 33.7 KB 33.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18661_fsc.xml emd_18661_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_18661.png emd_18661.png | 199.9 KB | ||
| Filedesc metadata |  emd-18661.cif.gz emd-18661.cif.gz | 9.4 KB | ||
| Others |  emd_18661_additional_1.map.gz emd_18661_additional_1.map.gz emd_18661_half_map_1.map.gz emd_18661_half_map_1.map.gz emd_18661_half_map_2.map.gz emd_18661_half_map_2.map.gz | 445.8 MB 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18661 http://ftp.pdbj.org/pub/emdb/structures/EMD-18661 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18661 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18661 | HTTPS FTP |
-Validation report
| Summary document |  emd_18661_validation.pdf.gz emd_18661_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18661_full_validation.pdf.gz emd_18661_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_18661_validation.xml.gz emd_18661_validation.xml.gz | 15.9 KB | Display | |
| Data in CIF |  emd_18661_validation.cif.gz emd_18661_validation.cif.gz | 21.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18661 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18661 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18661 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18661 | HTTPS FTP |
-Related structure data
| Related structure data |  8queMC  9rjsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18661.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18661.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PhiKZ nvRNAP DNA/RNA complex Postprocessed Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.92 Å | ||||||||||||||||||||||||||||||||||||
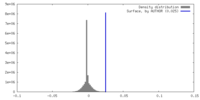
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: PhiKZ nvRNAP DNA/RNA complex Locally Filtered Map
| File | emd_18661_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PhiKZ nvRNAP DNA/RNA complex Locally Filtered Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
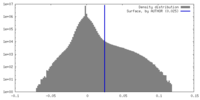
| Density Histograms |
-Half map: PhiKZ nvRNAP DNA/RNA complex Half Map 1
| File | emd_18661_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PhiKZ nvRNAP DNA/RNA complex Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
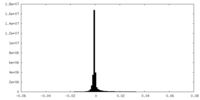
| Density Histograms |
-Half map: PhiKZ nvRNAP DNA/RNA complex Half Map 2
| File | emd_18661_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PhiKZ nvRNAP DNA/RNA complex Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
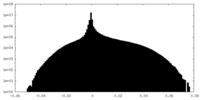
| Density Histograms |
- Sample components
Sample components
+Entire : PhiKZ non-virion RNA polymerase transcribing promoter P119L
+Supramolecule #1: PhiKZ non-virion RNA polymerase transcribing promoter P119L
+Macromolecule #1: PHIKZ055
+Macromolecule #2: DNA-directed RNA polymerase
+Macromolecule #3: PHIKZ074
+Macromolecule #4: PHIKZ123
+Macromolecule #8: PHIKZ068
+Macromolecule #5: DNA
+Macromolecule #6: DNA
+Macromolecule #9: DNA
+Macromolecule #7: RNA
+Macromolecule #10: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV Details: Settings: -6 blot force, 4 s waiting time and 0.5-1 s blotting time.. | ||||||||
| Details | PhiKZ nvRNAP complexes were mixed at 0.1 mg/mL final concentration with the P119L DNA/RNA template at a 1:1 ratio in the presence of 1 mM ATP, CTP and GTP. Reactions were incubated for 30 min at 37 C before use. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 3092 / Average electron dose: 51.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.9 µm / Nominal defocus min: 1.4000000000000001 µm / Nominal magnification: 15000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: CC | |||||||||||||||
| Output model |  PDB-8que: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)