[English] 日本語
 Yorodumi
Yorodumi- EMDB-18656: Mycobacterium smegnatis RNA polymerase transcription initiation c... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mycobacterium smegnatis RNA polymerase transcription initiation complex with SigmaA, RbpA, HelD and an upstream-fork promoter fragment | ||||||||||||||||||
 Map data Map data | Loc-Scale filtered | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | HelD / RNA polymerase / transcription initiation / Mycobacteria / TRANSCRIPTION / rifampicin resistance | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationrecombinational repair / 3'-5' DNA helicase activity / sigma factor activity / bacterial-type RNA polymerase core enzyme binding / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / protein dimerization activity ...recombinational repair / 3'-5' DNA helicase activity / sigma factor activity / bacterial-type RNA polymerase core enzyme binding / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / protein dimerization activity / hydrolase activity / response to antibiotic / DNA-templated transcription / positive regulation of DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / ATP binding / cytoplasm / cytosol Similarity search - Function | ||||||||||||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | ||||||||||||||||||
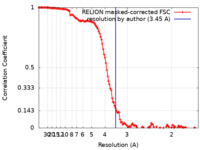
| Method | single particle reconstruction / cryo EM / Resolution: 3.45 Å | ||||||||||||||||||
 Authors Authors | Koval T / Krasny L / Dohnalek J / Kouba T | ||||||||||||||||||
| Funding support |  Czech Republic, European Union, 5 items Czech Republic, European Union, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Mycobacterial HelD connects RNA polymerase recycling with transcription initiation. Authors: Tomáš Kovaľ / Nabajyoti Borah / Petra Sudzinová / Barbora Brezovská / Hana Šanderová / Viola Vaňková Hausnerová / Alena Křenková / Martin Hubálek / Mária Trundová / Kristýna ...Authors: Tomáš Kovaľ / Nabajyoti Borah / Petra Sudzinová / Barbora Brezovská / Hana Šanderová / Viola Vaňková Hausnerová / Alena Křenková / Martin Hubálek / Mária Trundová / Kristýna Adámková / Jarmila Dušková / Marek Schwarz / Jana Wiedermannová / Jan Dohnálek / Libor Krásný / Tomáš Kouba /  Abstract: Mycobacterial HelD is a transcription factor that recycles stalled RNAP by dissociating it from nucleic acids and, if present, from the antibiotic rifampicin. The rescued RNAP, however, must ...Mycobacterial HelD is a transcription factor that recycles stalled RNAP by dissociating it from nucleic acids and, if present, from the antibiotic rifampicin. The rescued RNAP, however, must disengage from HelD to participate in subsequent rounds of transcription. The mechanism of release is unknown. We show that HelD from Mycobacterium smegmatis forms a complex with RNAP associated with the primary sigma factor σ and transcription factor RbpA but not CarD. We solve several structures of RNAP-σ-RbpA-HelD without and with promoter DNA. These snapshots capture HelD during transcription initiation, describing mechanistic aspects of HelD release from RNAP and its protective effect against rifampicin. Biochemical evidence supports these findings, defines the role of ATP binding and hydrolysis by HelD in the process, and confirms the rifampicin-protective effect of HelD. Collectively, these results show that when HelD is present during transcription initiation, the process is protected from rifampicin until the last possible moment. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18656.map.gz emd_18656.map.gz | 88.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18656-v30.xml emd-18656-v30.xml emd-18656.xml emd-18656.xml | 33.8 KB 33.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18656_fsc.xml emd_18656_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_18656.png emd_18656.png | 105.8 KB | ||
| Masks |  emd_18656_msk_1.map emd_18656_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18656.cif.gz emd-18656.cif.gz | 10.1 KB | ||
| Others |  emd_18656_additional_1.map.gz emd_18656_additional_1.map.gz emd_18656_half_map_1.map.gz emd_18656_half_map_1.map.gz emd_18656_half_map_2.map.gz emd_18656_half_map_2.map.gz | 165.7 MB 141.2 MB 140.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18656 http://ftp.pdbj.org/pub/emdb/structures/EMD-18656 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18656 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18656 | HTTPS FTP |
-Related structure data
| Related structure data |  8qu6MC  8q3iC  8qn8C  8qtiC  8r2mC  8r3mC  8r6pC  8r6rC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18656.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18656.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Loc-Scale filtered | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8336 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18656_msk_1.map emd_18656_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
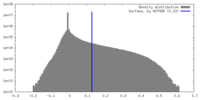
| Density Histograms |
-Additional map: RELION post-processed
| File | emd_18656_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION post-processed | ||||||||||||
| Projections & Slices |
| ||||||||||||
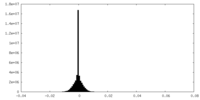
| Density Histograms |
-Half map: #1
| File | emd_18656_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18656_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Mycobacterium smegnatis RNA polymerase transcription initiation c...
+Supramolecule #1: Mycobacterium smegnatis RNA polymerase transcription initiation c...
+Macromolecule #1: DNA-directed RNA polymerase subunit alpha
+Macromolecule #2: DNA-directed RNA polymerase subunit beta
+Macromolecule #3: DNA-directed RNA polymerase subunit beta'
+Macromolecule #4: RNA polymerase-binding protein RbpA
+Macromolecule #5: DNA-directed RNA polymerase subunit omega
+Macromolecule #6: RNA polymerase sigma factor SigA
+Macromolecule #7: Helicase
+Macromolecule #8: DNA 22-MER
+Macromolecule #9: DNA 31-MER
+Macromolecule #10: ZINC ION
+Macromolecule #11: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 2.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.9 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)