+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | p97 (VCP) mutant - F539A with its adaptor | |||||||||
 Map data Map data | Post-processed map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Hexameric complex / ATPase / Unfoldase / Protein Quality Control / Segregase / CHAPERONE | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Arie M / Matzov D / Karmona R / Szenkier N / Stanhill A / Navon A | |||||||||
| Funding support |  Israel, 1 items Israel, 1 items
| |||||||||
 Citation Citation |  Journal: iScience / Year: 2024 Journal: iScience / Year: 2024Title: A non-symmetrical p97 conformation initiates a multistep recruitment of Ufd1/Npl4. Authors: Michal Arie / Donna Matzov / Rotem Karmona / Natalia Szenkier / Ariel Stanhill / Ami Navon /  Abstract: experiments and cryo-EM structures of p97 and its cofactor, Ufd1/Npl4 (UN), elucidated substrate processing. Yet, the structural transitions and the related ATPase cycle upon UN binding remain ... experiments and cryo-EM structures of p97 and its cofactor, Ufd1/Npl4 (UN), elucidated substrate processing. Yet, the structural transitions and the related ATPase cycle upon UN binding remain unresolved. We captured two discrete conformations: One in which D1 protomers are ATP bound, while the D2 subunits are in the ADP state, presumably required for substrate engagement with the D2 pore; and a heterologous nucleotide state within the D1 ring in which only two NTDs are in the "up" ATP state that favors UN binding. Further analysis suggests that initially, UN binds p97's non-symmetrical conformation, this association promotes a structural transition upon which five NTDs shift to an "up" state and are poised to bind ATP. The UBXL domain of Npl4 was captured bound to an NTD in the ADP state, demonstrating a conformation that may provide directionality to incoming substrate and introduce the flexibility needed for substrate processing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18517.map.gz emd_18517.map.gz | 23.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18517-v30.xml emd-18517-v30.xml emd-18517.xml emd-18517.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18517.png emd_18517.png | 166.7 KB | ||
| Filedesc metadata |  emd-18517.cif.gz emd-18517.cif.gz | 6.3 KB | ||
| Others |  emd_18517_additional_1.map.gz emd_18517_additional_1.map.gz emd_18517_half_map_1.map.gz emd_18517_half_map_1.map.gz emd_18517_half_map_2.map.gz emd_18517_half_map_2.map.gz | 139.9 MB 140.7 MB 140.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18517 http://ftp.pdbj.org/pub/emdb/structures/EMD-18517 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18517 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18517 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18517.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18517.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.793 Å | ||||||||||||||||||||||||||||||||||||
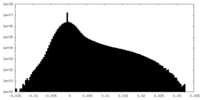
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_18517_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
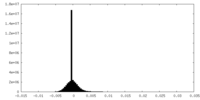
| Density Histograms |
-Half map: Half-map B
| File | emd_18517_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map A
| File | emd_18517_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
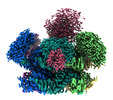
-Entire : Tertiary complex of the hexameric p97 F539A mutant with its adaptor
| Entire | Name: Tertiary complex of the hexameric p97 F539A mutant with its adaptor |
|---|---|
| Components |
|
-Supramolecule #1: Tertiary complex of the hexameric p97 F539A mutant with its adaptor
| Supramolecule | Name: Tertiary complex of the hexameric p97 F539A mutant with its adaptor type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: The mutant generated by the replacement of phenylalanine to alanine at position 539 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Transitional endoplasmic reticulum ATPase
| Macromolecule | Name: Transitional endoplasmic reticulum ATPase / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MASGADSKGD DLSTAILKQK NRPNRLIVDE AINEDNSVVS LSQPKMDELQ LFRGDTVLLK GKKRREAVCI VLSDDTCSDE KIRMNRVVR NNLRVRLGDV ISIQPCPDVK YGKRIHVLPI DDTVEGITGN LFEVYLKPYF LEAYRPIRKG DIFLVRGGMR A VEFKVVET ...String: MASGADSKGD DLSTAILKQK NRPNRLIVDE AINEDNSVVS LSQPKMDELQ LFRGDTVLLK GKKRREAVCI VLSDDTCSDE KIRMNRVVR NNLRVRLGDV ISIQPCPDVK YGKRIHVLPI DDTVEGITGN LFEVYLKPYF LEAYRPIRKG DIFLVRGGMR A VEFKVVET DPSPYCIVAP DTVIHCEGEP IKREDEEESL NEVGYDDIGG CRKQLAQIKE MVELPLRHPA LFKAIGVKPP RG ILLYGPP GTGKTLIARA VANETGAFFF LINGPEIMSK LAGESESNLR KAFEEAEKNA PAIIFIDELD AIAPKREKTH GEV ERRIVS QLLTLMDGLK QRAHVIVMAA TNRPNSIDPA LRRFGRFDRE VDIGIPDATG RLEILQIHTK NMKLADDVDL EQVA NETHG HVGADLAALC SEAALQAIRK KMDLIDLEDE TIDAEVMNSL AVTMDDFRWA LSQSNPSALR ETVVEVPQVT WEDIG GLED VKRELQELVQ YPVEHPDKFL KFGMTPSKGV LFYGPPGCGK TLLAKAIANE CQANAISIKG PELLTMWFGE SEANVR EIF DKARQAAPCV LFFDELDSIA KARGGNIGDG GGAADRVINQ ILTEMDGMST KKNVFIIGAT NRPDIIDPAI LRPGRLD QL IYIPLPDEKS RVAILKANLR KSPVAKDVDL EFLAKMTNGF SGADLTEICQ RACKLAIRES IESEIRRERE RQTNPSAM E VEEDDPVPEI RRDHFEEAMR FARRSVSDND IRKYEMFAQT LQQSRGFGSF RFPSGNQGGA GPSQGSGGGT GGSVYTEDN DDDLYG |
-Macromolecule #2: Nuclear protein localization protein 4 homolog
| Macromolecule | Name: Nuclear protein localization protein 4 homolog / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MAEGTIIRVQ SPDGVKRITA TKRETAATFL KKVAKEFGFQ NNGFSVYINR NKTGEITASS SKSLHLLKI KHGDLLFLFP SSLAGPSSEM ETSTSVGLKA FGAPHVVEDE IDQYLSKQDG K IYRSRDPQ LCRHGPLGKC VHCVPLEPFD EDYLNHLEPP VKHMSFHAYI ...String: MAEGTIIRVQ SPDGVKRITA TKRETAATFL KKVAKEFGFQ NNGFSVYINR NKTGEITASS SKSLHLLKI KHGDLLFLFP SSLAGPSSEM ETSTSVGLKA FGAPHVVEDE IDQYLSKQDG K IYRSRDPQ LCRHGPLGKC VHCVPLEPFD EDYLNHLEPP VKHMSFHAYI RKLTGGADKG KF VALENIS CKIKSGCEGH LPWPNGICTK CQPSAITLNR QKYRHVDNIM FENHTVADRF LDF WRKTGN QHFGYLYGRY TEHKDIPLGI RAEVAAIYEP PQIGTQNSLE LLEDPKAEVV DEIA SKLGL RKVGWIFTDL VSEDTRKGTV RYSRNKDTYF LSSEECITAG DFQNKHPNIC RLSPD GHFG SKFVTAVATG GPDNQVHFEG YQVSNQCMAL VRDECLLPCK DAPELGYAKE SSSEQY VPD VFYKDIDKFG NEITQLARPL PVEYLIIDIT TTFPKDPVYT FSISQNPFPI ENRDVLG ET QDFHSLATYL SQNTSSVFLD TISDFHLLLF LVTNEVMPLQ DSISLLLEAV RTRNEELA Q TWKKSEQWAT IEQLCSTVGV QLPGLHEFGA VGGSARAATS AMWACQHCTF MNQPGTGHC EMCSLPRT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: pre-blotting incubation time of 20 seconds and blot for 3.5 seconds with -1 blot force. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 11807 / Average electron dose: 43.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: Other / Chain - Initial model type: experimental model / Details: FROM EMDB |
|---|---|
| Refinement | Overall B value: 104 |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)