[English] 日本語
 Yorodumi
Yorodumi- EMDB-18505: Human 80S ribosome structure from pFIB-lamellae reprocessed with ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human 80S ribosome structure from pFIB-lamellae reprocessed with TomoBEAR | |||||||||||||||
 Map data Map data | Focused refinement of LSU, filtered to local resolution, calibrated apix 1.9 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | cryo-ET / tomography / StA / subtomogram averaging / tilt series / subnanometer resolution / pFIB / FIB-milling / plasma FIB / cell / RIBOSOME | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
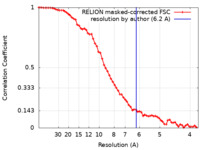
| Method | subtomogram averaging / cryo EM / Resolution: 6.2 Å | |||||||||||||||
 Authors Authors | Balyschew N / Yushkevich A / Mikirtumov V / Sanchez RM / Sprink T / Kudryashev M | |||||||||||||||
| Funding support |  Germany, 4 items Germany, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Streamlined structure determination by cryo-electron tomography and subtomogram averaging using TomoBEAR. Authors: Nikita Balyschew / Artsemi Yushkevich / Vasilii Mikirtumov / Ricardo M Sanchez / Thiemo Sprink / Mikhail Kudryashev /  Abstract: Structures of macromolecules in their native state provide unique unambiguous insights into their functions. Cryo-electron tomography combined with subtomogram averaging demonstrated the power to ...Structures of macromolecules in their native state provide unique unambiguous insights into their functions. Cryo-electron tomography combined with subtomogram averaging demonstrated the power to solve such structures in situ at resolutions in the range of 3 Angstrom for some macromolecules. In order to be applicable to the structural determination of the majority of macromolecules observable in cells in limited amounts, processing of tomographic data has to be performed in a high-throughput manner. Here we present TomoBEAR-a modular configurable workflow engine for streamlined processing of cryo-electron tomographic data for subtomogram averaging. TomoBEAR combines commonly used cryo-EM packages with reasonable presets to provide a transparent ("white box") approach for data management and processing. We demonstrate applications of TomoBEAR to two data sets of purified macromolecular targets, to an ion channel RyR1 in a membrane, and the tomograms of plasma FIB-milled lamellae and demonstrate the ability to produce high-resolution structures. TomoBEAR speeds up data processing, minimizes human interventions, and will help accelerate the adoption of in situ structural biology by cryo-ET. The source code and the documentation are freely available. #1:  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: Streamlined Structure Determination by Cryo-Electron Tomography and Subtomogram Averaging using TomoBEAR Authors: Balyschew N / Yushkevich A / Mikirtumov V / Sanchez RM / Sprink T / Kudryashev M | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18505.map.gz emd_18505.map.gz | 13.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18505-v30.xml emd-18505-v30.xml emd-18505.xml emd-18505.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18505_fsc.xml emd_18505_fsc.xml | 6.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_18505.png emd_18505.png | 147.1 KB | ||
| Masks |  emd_18505_msk_1.map emd_18505_msk_1.map | 22.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18505.cif.gz emd-18505.cif.gz | 4.8 KB | ||
| Others |  emd_18505_half_map_1.map.gz emd_18505_half_map_1.map.gz emd_18505_half_map_2.map.gz emd_18505_half_map_2.map.gz | 11 MB 11 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18505 http://ftp.pdbj.org/pub/emdb/structures/EMD-18505 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18505 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18505 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18505.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18505.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused refinement of LSU, filtered to local resolution, calibrated apix 1.9 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.9 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18505_msk_1.map emd_18505_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18505_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_18505_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 80S human ribosome
| Entire | Name: 80S human ribosome |
|---|---|
| Components |
|
-Supramolecule #1: 80S human ribosome
| Supramolecule | Name: 80S human ribosome / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 5.75 |
|---|---|
| Grid | Model: UltrAuFoil R2/2 / Material: GOLD / Mesh: 200 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV Details: 24 h post infection cells were plunge-frozen using the Vitrobot (Thermo Fisher) offsetting the blotting pad to favour back blotting. Just prior the PF, the cells media has been replaced with ...Details: 24 h post infection cells were plunge-frozen using the Vitrobot (Thermo Fisher) offsetting the blotting pad to favour back blotting. Just prior the PF, the cells media has been replaced with the complete media with 10% glycerol (v/v)... |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average electron dose: 5.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 5.0 µm / Calibrated defocus min: 2.5 µm / Calibrated magnification: 64000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)