+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the apo SPARTA (BabAgo/TIR-APAZ) complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Prokaryotic Argonaute / TIR domain / RNA binding protein / DNA binding protein / IMMUNE SYSTEM | |||||||||
| Biological species |  Bacillales bacterium (bacteria) Bacillales bacterium (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.57 Å | |||||||||
 Authors Authors | Finocchio G / Koopal B / Potocnik A / Heijstek C / Jinek M / Swarts D | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2024 Journal: Nucleic Acids Res / Year: 2024Title: Target DNA-dependent activation mechanism of the prokaryotic immune system SPARTA. Authors: Giada Finocchio / Balwina Koopal / Ana Potocnik / Clint Heijstek / Adrie H Westphal / Martin Jinek / Daan C Swarts /   Abstract: In both prokaryotic and eukaryotic innate immune systems, TIR domains function as NADases that degrade the key metabolite NAD+ or generate signaling molecules. Catalytic activation of TIR domains ...In both prokaryotic and eukaryotic innate immune systems, TIR domains function as NADases that degrade the key metabolite NAD+ or generate signaling molecules. Catalytic activation of TIR domains requires oligomerization, but how this is achieved varies in distinct immune systems. In the Short prokaryotic Argonaute (pAgo)/TIR-APAZ (SPARTA) immune system, TIR NADase activity is triggered upon guide RNA-mediated recognition of invading DNA by an unknown mechanism. Here, we describe cryo-EM structures of SPARTA in the inactive monomeric and target DNA-activated tetrameric states. The monomeric SPARTA structure reveals that in the absence of target DNA, a C-terminal tail of TIR-APAZ occupies the nucleic acid binding cleft formed by the pAgo and TIR-APAZ subunits, inhibiting SPARTA activation. In the active tetrameric SPARTA complex, guide RNA-mediated target DNA binding displaces the C-terminal tail and induces conformational changes in pAgo that facilitate SPARTA-SPARTA dimerization. Concurrent release and rotation of one TIR domain allow it to form a composite NADase catalytic site with the other TIR domain within the dimer, and generate a self-complementary interface that mediates cooperative tetramerization. Combined, this study provides critical insights into the structural architecture of SPARTA and the molecular mechanism underlying target DNA-dependent oligomerization and catalytic activation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18486.map.gz emd_18486.map.gz | 89 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18486-v30.xml emd-18486-v30.xml emd-18486.xml emd-18486.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_18486.png emd_18486.png | 49 KB | ||
| Filedesc metadata |  emd-18486.cif.gz emd-18486.cif.gz | 6.4 KB | ||
| Others |  emd_18486_half_map_1.map.gz emd_18486_half_map_1.map.gz emd_18486_half_map_2.map.gz emd_18486_half_map_2.map.gz | 165.1 MB 165.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18486 http://ftp.pdbj.org/pub/emdb/structures/EMD-18486 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18486 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18486 | HTTPS FTP |
-Validation report
| Summary document |  emd_18486_validation.pdf.gz emd_18486_validation.pdf.gz | 810.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18486_full_validation.pdf.gz emd_18486_full_validation.pdf.gz | 809.8 KB | Display | |
| Data in XML |  emd_18486_validation.xml.gz emd_18486_validation.xml.gz | 14.9 KB | Display | |
| Data in CIF |  emd_18486_validation.cif.gz emd_18486_validation.cif.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18486 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18486 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18486 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18486 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18486.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18486.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||
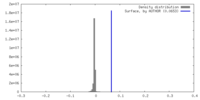
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_18486_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
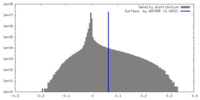
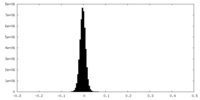
| Density Histograms |
-Half map: #1
| File | emd_18486_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
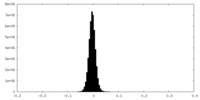
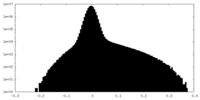
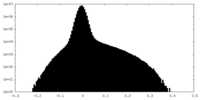
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of BabAgo with BabTIR-APAZ
| Entire | Name: Complex of BabAgo with BabTIR-APAZ |
|---|---|
| Components |
|
-Supramolecule #1: Complex of BabAgo with BabTIR-APAZ
| Supramolecule | Name: Complex of BabAgo with BabTIR-APAZ / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Bacillales bacterium (bacteria) Bacillales bacterium (bacteria) |
-Macromolecule #1: Short prokaryotic Argonaute
| Macromolecule | Name: Short prokaryotic Argonaute / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacillales bacterium (bacteria) Bacillales bacterium (bacteria) |
| Molecular weight | Theoretical: 57.911199 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKELIYIHEP NILFANGQKC ADPRDGLALF GPFTKIYGIK SGVVGTQYGL SIFKNYINHI QKPIYNANNI TRPMFPGFEA VFGCKWDAD NVVFKEVTKE EIEKILYTES NHKRTYDLVS LFINKIITAN KNEDEKVDVW FLVIPDEIYQ YCRPNSVLPK D LVQTKSLI ...String: MKELIYIHEP NILFANGQKC ADPRDGLALF GPFTKIYGIK SGVVGTQYGL SIFKNYINHI QKPIYNANNI TRPMFPGFEA VFGCKWDAD NVVFKEVTKE EIEKILYTES NHKRTYDLVS LFINKIITAN KNEDEKVDVW FLVIPDEIYQ YCRPNSVLPK D LVQTKSLI TKSKAKSFRY EPTLFENINK ELKEQEKEAI TYNYDAQFHD QLKARLLEHT IPTQILREST LAWRDFKNKF GA PKRDFSK IEGHLAWTIS TAAFYKAGGK PWKLSDIRSG VCYLGLVYKQ IEKSSNPKNA CCAAQMFLDN GDGTVFKGEV GPW YNQEKH EFHLNPKEAK ALLTQALNSY KEQNGVFPKE IFIHAKTKFN GQEWNAFQEV TPEGTNLVGV TITKTKPLKL FKSE GNYPI MRGNAFIVNE RSAFLWTVGY VPKTESTLSM EVPNPIFIEI NKGEADIEQV LKDVLALTKL NYNACIYADG VPVTL RFAD KIGEILTAST ELKAPPLAFK YYI |
-Macromolecule #2: Toll/interleukin-1 receptor domain-containing protein
| Macromolecule | Name: Toll/interleukin-1 receptor domain-containing protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacillales bacterium (bacteria) Bacillales bacterium (bacteria) |
| Molecular weight | Theoretical: 53.052355 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNARNKIFIS HAAPDDNDFT KWLALKLIAL GYEVWCDVLF LDKGADFWKV IDKEIREGAI KFLLATSEIA IKRDGVLKEI AVAEKVKKQ LKDDNFIIPL IIDENLSYDD LPPEIIRLNA VDFKKSWAVG LQDLLKALDD QKVEKNSPDP DKSNALYQQI F LHNKGIIE ...String: SNARNKIFIS HAAPDDNDFT KWLALKLIAL GYEVWCDVLF LDKGADFWKV IDKEIREGAI KFLLATSEIA IKRDGVLKEI AVAEKVKKQ LKDDNFIIPL IIDENLSYDD LPPEIIRLNA VDFKKSWAVG LQDLLKALDD QKVEKNSPDP DKSNALYQQI F LHNKGIIE REEIYDSNWF SILSFPKELR FHDYEKLMPK GFDVRELTYP AVRYKNYLCT FAWEYDFMHQ LPKTETYNSS QT IRIPTEE ILSGKYDSPF IGNFECQRLI VQLLNKAFEL RMKEKGVREY PMSNKMGYWF EKGKLEKDKF NKVLLVGKQK DKH WHFGIS AAGKLYPFPV LMISSHIFFT KDGKELIESK KIQHAARRRQ GKNWWNDDWR NKLLAFVKYL SDDENSFYLE VGSE EKIYI SNEPVQFVGK VSYNMPEKNN LKDEAEISDL NDLNEFDGEI FEETDSE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 58.005 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)