[English] 日本語
 Yorodumi
Yorodumi- EMDB-1841: Structural basis for the subunit assembly of the anaphase promoti... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1841 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural basis for the subunit assembly of the anaphase promoting complex | |||||||||
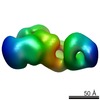
 Map data Map data | Anaphase promoting complex TPR6 subcomplex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | anaphase promoting complex / cyclosome / APC / APC/C / cell cycle / D-box / KEN-box / co-activator / Cdh1 / tetraticopeptide repeats / TPR / ubiquitylation / cyclin | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining | |||||||||
 Authors Authors | Schreiber A / Stengel F / Zhang Z / Enchev RE / Kong E / Morris EP / Robinson CV / daFonseca PCA / Barford D | |||||||||
 Citation Citation |  Journal: Nature / Year: 2011 Journal: Nature / Year: 2011Title: Structural basis for the subunit assembly of the anaphase-promoting complex. Authors: Anne Schreiber / Florian Stengel / Ziguo Zhang / Radoslav I Enchev / Eric H Kong / Edward P Morris / Carol V Robinson / Paula C A da Fonseca / David Barford /  Abstract: The anaphase-promoting complex or cyclosome (APC/C) is an unusually large E3 ubiquitin ligase responsible for regulating defined cell cycle transitions. Information on how its 13 constituent proteins ...The anaphase-promoting complex or cyclosome (APC/C) is an unusually large E3 ubiquitin ligase responsible for regulating defined cell cycle transitions. Information on how its 13 constituent proteins are assembled, and how they interact with co-activators, substrates and regulatory proteins is limited. Here, we describe a recombinant expression system that allows the reconstitution of holo APC/C and its sub-complexes that, when combined with electron microscopy, mass spectrometry and docking of crystallographic and homology-derived coordinates, provides a precise definition of the organization and structure of all essential APC/C subunits, resulting in a pseudo-atomic model for 70% of the APC/C. A lattice-like appearance of the APC/C is generated by multiple repeat motifs of most APC/C subunits. Three conserved tetratricopeptide repeat (TPR) subunits (Cdc16, Cdc23 and Cdc27) share related superhelical homo-dimeric architectures that assemble to generate a quasi-symmetrical structure. Our structure explains how this TPR sub-complex, together with additional scaffolding subunits (Apc1, Apc4 and Apc5), coordinate the juxtaposition of the catalytic and substrate recognition module (Apc2, Apc11 and Apc10 (also known as Doc1)), and TPR-phosphorylation sites, relative to co-activator, regulatory proteins and substrates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1841.map.gz emd_1841.map.gz | 10 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1841-v30.xml emd-1841-v30.xml emd-1841.xml emd-1841.xml | 7.9 KB 7.9 KB | Display Display |  EMDB header EMDB header |
| Images |  figure_EBI1841.tif figure_EBI1841.tif | 130.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1841 http://ftp.pdbj.org/pub/emdb/structures/EMD-1841 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1841 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1841 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1841.map.gz / Format: CCP4 / Size: 10.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1841.map.gz / Format: CCP4 / Size: 10.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Anaphase promoting complex TPR6 subcomplex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.47 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Anaphase promoting complex TPR6 subcomplex
| Entire | Name: Anaphase promoting complex TPR6 subcomplex |
|---|---|
| Components |
|
-Supramolecule #1000: Anaphase promoting complex TPR6 subcomplex
| Supramolecule | Name: Anaphase promoting complex TPR6 subcomplex / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Macromolecule #1: Anaphase promoting complex TPR6 subcomplex
| Macromolecule | Name: Anaphase promoting complex TPR6 subcomplex / type: protein_or_peptide / ID: 1 / Name.synonym: Anaphase promoting complex TPR6 subcomplex / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism: Baculovirus/Insect Cells |
-Macromolecule #2: Cdh1
| Macromolecule | Name: Cdh1 / type: protein_or_peptide / ID: 2 / Name.synonym: Cdh1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism: Baculovirus/Insect cells |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Staining | Type: NEGATIVE / Details: Grids were stained using 2% w/v uranyl acetate |
|---|---|
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Details | Sample stained using uranyl acetate and imaged at room temperature |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Average electron dose: 100 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Room temperature side entry / Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) |
|---|
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















