+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | UFL1 E3 ligase bound 60S ribosome | |||||||||||||||
 Map data Map data | DeepEMhancer map. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | UFM1 / Ligase / Ribosome / Complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of I-kappaB phosphorylation / UFM1 ligase activity / UFM1-modified protein reader activity / positive regulation of reticulophagy / regulation of phosphatase activity / apoptotic nuclear changes / definitive erythrocyte differentiation / negative regulation of protein serine/threonine kinase activity / UFM1 transferase activity / positive regulation of protein localization to endoplasmic reticulum ...positive regulation of I-kappaB phosphorylation / UFM1 ligase activity / UFM1-modified protein reader activity / positive regulation of reticulophagy / regulation of phosphatase activity / apoptotic nuclear changes / definitive erythrocyte differentiation / negative regulation of protein serine/threonine kinase activity / UFM1 transferase activity / positive regulation of protein localization to endoplasmic reticulum / positive regulation of proteolysis involved in protein catabolic process / protein K69-linked ufmylation / negative regulation of protein kinase activity by regulation of protein phosphorylation / protein ufmylation / positive regulation of plasma cell differentiation / negative regulation of IRE1-mediated unfolded protein response / regulation of proteasomal ubiquitin-dependent protein catabolic process / positive regulation of cell cycle G1/S phase transition / protein localization to endoplasmic reticulum / negative regulation of T cell mediated immune response to tumor cell / negative regulation of T cell activation / regulation of intracellular estrogen receptor signaling pathway / positive regulation of proteasomal protein catabolic process / mitotic G2/M transition checkpoint / regulation of cyclin-dependent protein serine/threonine kinase activity / cartilage development / ribosome disassembly / Transferases; Acyltransferases; Aminoacyltransferases / mitogen-activated protein kinase binding / regulation of canonical NF-kappaB signal transduction / reticulophagy / regulation of neuron differentiation / response to L-glutamate / negative regulation of NF-kappaB transcription factor activity / mitotic G2 DNA damage checkpoint signaling / negative regulation of PERK-mediated unfolded protein response / negative regulation of protein phosphorylation / negative regulation of MAP kinase activity / ubiquitin-like protein ligase binding / Peptide chain elongation / Selenocysteine synthesis / Formation of a pool of free 40S subunits / Eukaryotic Translation Termination / Response of EIF2AK4 (GCN2) to amino acid deficiency / positive regulation of signal transduction by p53 class mediator / SRP-dependent cotranslational protein targeting to membrane / RHOA GTPase cycle / Viral mRNA Translation / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / hematopoietic stem cell differentiation / GTP hydrolysis and joining of the 60S ribosomal subunit / NF-kappaB binding / L13a-mediated translational silencing of Ceruloplasmin expression / ubiquitin-like ligase-substrate adaptor activity / Major pathway of rRNA processing in the nucleolus and cytosol / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / endoplasmic reticulum unfolded protein response / positive regulation of protein ubiquitination / positive regulation of glial cell proliferation / MDM2/MDM4 family protein binding / negative regulation of proteasomal ubiquitin-dependent protein catabolic process / negative regulation of protein ubiquitination / positive regulation of autophagy / cytosolic ribosome / rescue of stalled ribosome / endomembrane system / cyclin binding / regulation of mitotic cell cycle / response to endoplasmic reticulum stress / DNA damage checkpoint signaling / erythrocyte differentiation / liver development / positive regulation of NF-kappaB transcription factor activity / Regulation of expression of SLITs and ROBOs / negative regulation of protein catabolic process / brain development / regulation of protein stability / positive regulation of protein localization to nucleus / osteoblast differentiation / Antigen processing: Ubiquitination & Proteasome degradation / positive regulation of proteasomal ubiquitin-dependent protein catabolic process / site of double-strand break / regulation of protein localization / regulation of inflammatory response / response to ethanol / cytosolic large ribosomal subunit / mitochondrial outer membrane / cytoplasmic translation / neuron projection / positive regulation of canonical NF-kappaB signal transduction / cell population proliferation / postsynaptic density / protein stabilization / positive regulation of cell migration / structural constituent of ribosome / translation / negative regulation of gene expression / focal adhesion / DNA repair / intracellular membrane-bounded organelle Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
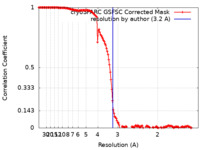
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||||||||
 Authors Authors | Makhlouf L / Zeqiraj E / Kulathu Y | |||||||||||||||
| Funding support |  United Kingdom, European Union, 4 items United Kingdom, European Union, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons. Authors: Linda Makhlouf / Joshua J Peter / Helge M Magnussen / Rohan Thakur / David Millrine / Thomas C Minshull / Grace Harrison / Joby Varghese / Frederic Lamoliatte / Martina Foglizzo / Thomas ...Authors: Linda Makhlouf / Joshua J Peter / Helge M Magnussen / Rohan Thakur / David Millrine / Thomas C Minshull / Grace Harrison / Joby Varghese / Frederic Lamoliatte / Martina Foglizzo / Thomas Macartney / Antonio N Calabrese / Elton Zeqiraj / Yogesh Kulathu /  Abstract: Stalled ribosomes at the endoplasmic reticulum (ER) are covalently modified with the ubiquitin-like protein UFM1 on the 60S ribosomal subunit protein RPL26 (also known as uL24). This modification, ...Stalled ribosomes at the endoplasmic reticulum (ER) are covalently modified with the ubiquitin-like protein UFM1 on the 60S ribosomal subunit protein RPL26 (also known as uL24). This modification, which is known as UFMylation, is orchestrated by the UFM1 ribosome E3 ligase (UREL) complex, comprising UFL1, UFBP1 and CDK5RAP3 (ref. ). However, the catalytic mechanism of UREL and the functional consequences of UFMylation are unclear. Here we present cryo-electron microscopy structures of UREL bound to 60S ribosomes, revealing the basis of its substrate specificity. UREL wraps around the 60S subunit to form a C-shaped clamp architecture that blocks the tRNA-binding sites at one end, and the peptide exit tunnel at the other. A UFL1 loop inserts into and remodels the peptidyl transferase centre. These features of UREL suggest a crucial function for UFMylation in the release and recycling of stalled or terminated ribosomes from the ER membrane. In the absence of functional UREL, 60S-SEC61 translocon complexes accumulate at the ER membrane, demonstrating that UFMylation is necessary for releasing SEC61 from 60S subunits. Notably, this release is facilitated by a functional switch of UREL from a 'writer' to a 'reader' module that recognizes its product-UFMylated 60S ribosomes. Collectively, we identify a fundamental role for UREL in dissociating 60S subunits from the SEC61 translocon and the basis for UFMylation in regulating protein homeostasis at the ER. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18381.map.gz emd_18381.map.gz | 270.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18381-v30.xml emd-18381-v30.xml emd-18381.xml emd-18381.xml | 28 KB 28 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18381_fsc.xml emd_18381_fsc.xml | 14.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_18381.png emd_18381.png | 44.9 KB | ||
| Masks |  emd_18381_msk_1.map emd_18381_msk_1.map | 775.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18381.cif.gz emd-18381.cif.gz | 8.1 KB | ||
| Others |  emd_18381_additional_1.map.gz emd_18381_additional_1.map.gz emd_18381_half_map_1.map.gz emd_18381_half_map_1.map.gz emd_18381_half_map_2.map.gz emd_18381_half_map_2.map.gz | 287 MB 9 MB 9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18381 http://ftp.pdbj.org/pub/emdb/structures/EMD-18381 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18381 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18381 | HTTPS FTP |
-Validation report
| Summary document |  emd_18381_validation.pdf.gz emd_18381_validation.pdf.gz | 446.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18381_full_validation.pdf.gz emd_18381_full_validation.pdf.gz | 446 KB | Display | |
| Data in XML |  emd_18381_validation.xml.gz emd_18381_validation.xml.gz | 27 KB | Display | |
| Data in CIF |  emd_18381_validation.cif.gz emd_18381_validation.cif.gz | 34.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18381 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18381 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18381 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18381 | HTTPS FTP |
-Related structure data
| Related structure data |  8qfcMC  8bzrC  8c0dC  8qfdC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18381.map.gz / Format: CCP4 / Size: 775.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18381.map.gz / Format: CCP4 / Size: 775.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer map. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.74 Å | ||||||||||||||||||||||||||||||||||||
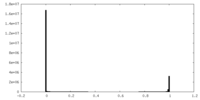
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18381_msk_1.map emd_18381_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
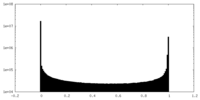
| Density Histograms |
-Additional map: Raw map.
| File | emd_18381_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
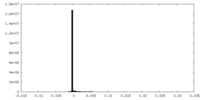
| Density Histograms |
-Half map: #2
| File | emd_18381_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
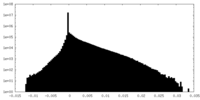
| Density Histograms |
-Half map: #1
| File | emd_18381_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : UFM1 ribosome E3 ligase complex bound to 60S ribosome
| Entire | Name: UFM1 ribosome E3 ligase complex bound to 60S ribosome |
|---|---|
| Components |
|
-Supramolecule #1: UFM1 ribosome E3 ligase complex bound to 60S ribosome
| Supramolecule | Name: UFM1 ribosome E3 ligase complex bound to 60S ribosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: 60S ribosomal protein L10a
| Macromolecule | Name: 60S ribosomal protein L10a / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.879422 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSKVSRDTL YEAVREVLHG NQRKRRKFLE TVELQISLKN YDPQKDKRFS GTVRLKSTPR PKFSVCVLGD QQHCDEAKAV DIPHMDIEA LKKLNKNKKL VKKLAKKYDA FLASESLIKQ IPRILGPGLN KAGKFPSLLT HNENMVAKVD EVKSTIKFQM K KVLCLAVA ...String: MSSKVSRDTL YEAVREVLHG NQRKRRKFLE TVELQISLKN YDPQKDKRFS GTVRLKSTPR PKFSVCVLGD QQHCDEAKAV DIPHMDIEA LKKLNKNKKL VKKLAKKYDA FLASESLIKQ IPRILGPGLN KAGKFPSLLT HNENMVAKVD EVKSTIKFQM K KVLCLAVA VGHVKMTDDE LVYNIHLAVN FLVSLLKKNW QNVRALYIKS TMGKPQRLY UniProtKB: Large ribosomal subunit protein uL1 |
-Macromolecule #2: E3 UFM1-protein ligase 1
| Macromolecule | Name: E3 UFM1-protein ligase 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Transferases; Acyltransferases; Aminoacyltransferases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 91.591234 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHEN LYFQGMADAW EEIRRLAADF QRAQFAEATQ RLSERNCIEI VNKLIAQKQL EVVHTLDGKE YITPAQISKE MRDELHVRG GRVNIVDLQQ VINVDLIHIE NRIGDIIKSE KHVQLVLGQL IDENYLDRLA EEVNDKLQES GQVTISELCK T YDLPGNFL ...String: MGHHHHHHEN LYFQGMADAW EEIRRLAADF QRAQFAEATQ RLSERNCIEI VNKLIAQKQL EVVHTLDGKE YITPAQISKE MRDELHVRG GRVNIVDLQQ VINVDLIHIE NRIGDIIKSE KHVQLVLGQL IDENYLDRLA EEVNDKLQES GQVTISELCK T YDLPGNFL TQALTQRLGR IISGHIDLDN RGVIFTEAFV ARHKARIRGL FSAITRPTAV NSLISKYGFQ EQLLYSVLEE LV NSGRLRG TVVGGRQDKA VFVPDIYSRT QSTWVDSFFR QNGYLEFDAL SRLGIPDAVS YIKKRYKTTQ LLFLKAACVG QGL VDQVEA SVEEAISSGT WVDIAPLLPT SLSVEDAAIL LQQVMRAFSK QASTVVFSDT VVVSEKFIND CTELFRELMH QKAE KEMKN NPVHLITEED LKQISTLESV STSKKDKKDE RRRKATEGSG SMRGGGGGNA REYKIKKVKK KGRKDDDSDD ESQSS HTGK KKPEISFMFQ DEIEDFLRKH IQDAPEEFIS ELAEYLIKPL NKTYLEVVRS VFMSSTTSAS GTGRKRTIKD LQEEVS NLY NNIRLFEKGM KFFADDTQAA LTKHLLKSVC TDITNLIFNF LASDLMMAVD DPAAITSEIR KKILSKLSEE TKVALTK LH NSLNEKSIED FISCLDSAAE ACDIMVKRGD KKRERQILFQ HRQALAEQLK VTEDPALILH LTSVLLFQFS THSMLHAP G RCVPQIIAFL NSKIPEDQHA LLVKYQGLVV KQLVSQSKKT GQGDYPLNNE LDKEQEDVAS TTRKELQELS SSIKDLVLK SRKSSVTEE UniProtKB: E3 UFM1-protein ligase 1 |
-Macromolecule #3: CDK5 regulatory subunit-associated protein 3
| Macromolecule | Name: CDK5 regulatory subunit-associated protein 3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.458938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPLVDMEDHQ HVPIDIQTSK LLDWLVDRRH CSLKWQSLVL TIREKINAAI QDMPESEEIA QLLSGSYIHY FHCLRILDLL KGTEASTKN IFGRYSSQRM KDWQEIIALY EKDNTYLVEL SSLLVRNVNY EIPSLKKQIA KCQQLQQEYS RKEEECQAGA A EMREQFYH ...String: GPLVDMEDHQ HVPIDIQTSK LLDWLVDRRH CSLKWQSLVL TIREKINAAI QDMPESEEIA QLLSGSYIHY FHCLRILDLL KGTEASTKN IFGRYSSQRM KDWQEIIALY EKDNTYLVEL SSLLVRNVNY EIPSLKKQIA KCQQLQQEYS RKEEECQAGA A EMREQFYH SCKQYGITGE NVRGELLALV KDLPSQLAEI GAAAQQSLGE AIDVYQASVG FVCESPTEQV LPMLRFVQKR GN STVYEWR TGTEPSVVER PHLEELPEQV AEDAIDWGDF GVEAVSEGTD SGISAEAAGI DWGIFPESDS KDPGGDGIDW GDD AVALQI TVLEAGTQAP EGVARGPDAL TLLEYTETRN QFLDELMELE IFLAQRAVEL SEEADVLSVS QFQLAPAILQ GQTK EKMVT MVSVLEDLIG KLTSLQLQHL FMILASPRYV DRVTEFLQQK LKQSQLLALK KELMVQKQQE ALEEQAALEP KLDLL LEKT KELQKLIEAD ISKRYSGRPV NLMGTSL UniProtKB: CDK5 regulatory subunit-associated protein 3 |
-Macromolecule #4: DDRGK domain-containing protein 1
| Macromolecule | Name: DDRGK domain-containing protein 1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.644445 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MWSHPQFEKL EVLFQGPASA GQEPLHNEEL AGAGRVAQPG PLEPEEPRAG GRPRRRRDLG SRLQAQRRAQ RVAWAEADEN EEEAVILAQ EEEGVEKPAE THLSGKIGAK KLRKLEEKQA RKAQREAEEA EREERKRLES QREAEWKKEE ERLRLEEEQK E EEERKARE ...String: MWSHPQFEKL EVLFQGPASA GQEPLHNEEL AGAGRVAQPG PLEPEEPRAG GRPRRRRDLG SRLQAQRRAQ RVAWAEADEN EEEAVILAQ EEEGVEKPAE THLSGKIGAK KLRKLEEKQA RKAQREAEEA EREERKRLES QREAEWKKEE ERLRLEEEQK E EEERKARE EQAQREHEEY LKLKEAFVVE EEGVGETMTE EQSQSFLTEF INYIKQSKVV LLEDLASQVG LRTQDTINRI QD LLAEGTI TGVIDDRGKF IYITPEELAA VANFIRQRGR VSIAELAQAS NSLIAWGRES PAQAPA UniProtKB: DDRGK domain-containing protein 1 |
-Macromolecule #5: Ubiquitin-fold modifier 1
| Macromolecule | Name: Ubiquitin-fold modifier 1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.236628 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPGSMSKVSF KITLTSDPRL PYKVLSVPES TPFTAVLKFA AEEFKVPAAT SAIITNDGIG INPAQTAGNV FLKHGSELRI IPRDRVG UniProtKB: Ubiquitin-fold modifier 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7.7 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 25 mM HEPES pH 7.5, 50 mM KCl, 5 mM MgCl2, 2 mM DTT |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 59394 / Average exposure time: 2.67 sec. / Average electron dose: 33.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.2 µm / Nominal magnification: 165000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)