+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tau - CTE-LIA4 (tau intermediate amyloid) | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Amyloid / tau / PROTEIN FIBRIL | |||||||||
| Function / homology |  Function and homology information Function and homology informationplus-end-directed organelle transport along microtubule / histone-dependent DNA binding / negative regulation of establishment of protein localization to mitochondrion / neurofibrillary tangle / microtubule lateral binding / axonal transport / main axon / phosphatidylinositol bisphosphate binding / regulation of long-term synaptic depression / tubulin complex ...plus-end-directed organelle transport along microtubule / histone-dependent DNA binding / negative regulation of establishment of protein localization to mitochondrion / neurofibrillary tangle / microtubule lateral binding / axonal transport / main axon / phosphatidylinositol bisphosphate binding / regulation of long-term synaptic depression / tubulin complex / positive regulation of protein localization to synapse / negative regulation of tubulin deacetylation / generation of neurons / regulation of chromosome organization / rRNA metabolic process / axonal transport of mitochondrion / regulation of mitochondrial fission / axon development / central nervous system neuron development / intracellular distribution of mitochondria / regulation of microtubule polymerization / microtubule polymerization / lipoprotein particle binding / minor groove of adenine-thymine-rich DNA binding / dynactin binding / negative regulation of mitochondrial membrane potential / apolipoprotein binding / axolemma / protein polymerization / glial cell projection / negative regulation of mitochondrial fission / Caspase-mediated cleavage of cytoskeletal proteins / regulation of microtubule polymerization or depolymerization / positive regulation of axon extension / neurofibrillary tangle assembly / Activation of AMPK downstream of NMDARs / regulation of cellular response to heat / synapse assembly / supramolecular fiber organization / positive regulation of protein localization / regulation of calcium-mediated signaling / somatodendritic compartment / cellular response to brain-derived neurotrophic factor stimulus / cytoplasmic microtubule organization / positive regulation of microtubule polymerization / axon cytoplasm / stress granule assembly / phosphatidylinositol binding / regulation of microtubule cytoskeleton organization / nuclear periphery / protein phosphatase 2A binding / positive regulation of superoxide anion generation / cellular response to reactive oxygen species / astrocyte activation / Hsp90 protein binding / microglial cell activation / cellular response to nerve growth factor stimulus / response to lead ion / synapse organization / PKR-mediated signaling / protein homooligomerization / regulation of synaptic plasticity / SH3 domain binding / memory / microtubule cytoskeleton organization / cytoplasmic ribonucleoprotein granule / neuron projection development / cell-cell signaling / single-stranded DNA binding / protein-folding chaperone binding / actin binding / cellular response to heat / microtubule cytoskeleton / cell body / growth cone / double-stranded DNA binding / microtubule binding / protein-macromolecule adaptor activity / dendritic spine / sequence-specific DNA binding / microtubule / amyloid fibril formation / learning or memory / neuron projection / regulation of autophagy / membrane raft / axon / negative regulation of gene expression / neuronal cell body / dendrite / DNA damage response / protein kinase binding / enzyme binding / mitochondrion / DNA binding / RNA binding / extracellular region / identical protein binding / nucleus / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
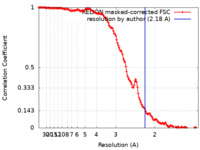
| Method | helical reconstruction / cryo EM / Resolution: 2.18 Å | |||||||||
 Authors Authors | Lovestam S / Li D / Scheres SHW / Goedert M | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Disease-specific tau filaments assemble via polymorphic intermediates. Authors: Sofia Lövestam / David Li / Jane L Wagstaff / Abhay Kotecha / Dari Kimanius / Stephen H McLaughlin / Alexey G Murzin / Stefan M V Freund / Michel Goedert / Sjors H W Scheres /   Abstract: Intermediate species in the assembly of amyloid filaments are believed to play a central role in neurodegenerative diseases and may constitute important targets for therapeutic intervention. However, ...Intermediate species in the assembly of amyloid filaments are believed to play a central role in neurodegenerative diseases and may constitute important targets for therapeutic intervention. However, structural information about intermediate species has been scarce and the molecular mechanisms by which amyloids assemble remain largely unknown. Here we use time-resolved cryogenic electron microscopy to study the in vitro assembly of recombinant truncated tau (amino acid residues 297-391) into paired helical filaments of Alzheimer's disease or into filaments of chronic traumatic encephalopathy. We report the formation of a shared first intermediate amyloid filament, with an ordered core comprising residues 302-316. Nuclear magnetic resonance indicates that the same residues adopt rigid, β-strand-like conformations in monomeric tau. At later time points, the first intermediate amyloid disappears and we observe many different intermediate amyloid filaments, with structures that depend on the reaction conditions. At the end of both assembly reactions, most intermediate amyloids disappear and filaments with the same ordered cores as those from human brains remain. Our results provide structural insights into the processes of primary and secondary nucleation of amyloid assembly, with implications for the design of new therapies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18281.map.gz emd_18281.map.gz | 57.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18281-v30.xml emd-18281-v30.xml emd-18281.xml emd-18281.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18281_fsc.xml emd_18281_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_18281.png emd_18281.png | 38.6 KB | ||
| Filedesc metadata |  emd-18281.cif.gz emd-18281.cif.gz | 5.3 KB | ||
| Others |  emd_18281_additional_1.map.gz emd_18281_additional_1.map.gz emd_18281_half_map_1.map.gz emd_18281_half_map_1.map.gz emd_18281_half_map_2.map.gz emd_18281_half_map_2.map.gz | 171.1 MB 171.2 MB 171.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18281 http://ftp.pdbj.org/pub/emdb/structures/EMD-18281 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18281 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18281 | HTTPS FTP |
-Related structure data
| Related structure data |  8q9hMC  8ppoC  8q27C  8q2jC  8q2kC  8q2lC  8q7fC  8q7lC  8q7mC  8q7pC  8q7tC  8q88C  8q8cC  8q8dC  8q8eC  8q8fC  8q8lC  8q8mC  8q8rC  8q8sC  8q8uC  8q8vC  8q8wC  8q8xC  8q8yC  8q8zC  8q97C  8q98C  8q99C  8q9aC  8q9bC  8q9cC  8q9dC  8q9eC  8q9fC  8q9gC  8q9iC  8q9jC  8q9kC  8q9lC  8q9mC  8q9oC  8qcpC  8qcrC  8qjjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18281.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18281.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
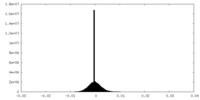
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.727 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_18281_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
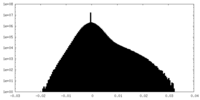
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_18281_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_18281_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Amyloid
| Entire | Name: Amyloid |
|---|---|
| Components |
|
-Supramolecule #1: Amyloid
| Supramolecule | Name: Amyloid / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Isoform Tau-D of Microtubule-associated protein tau
| Macromolecule | Name: Isoform Tau-D of Microtubule-associated protein tau / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.002773 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAEPRQEFEV MEDHAGTYGL GDRKDQGGYT MHQDQEGDTD AGLKEEAGIG DTPSLEDEAA GHVTQARMVS KSKDGTGSDD KKAKGADGK TKIATPRGAA PPGQKGQANA TRIPAKTPPA PKTPPSSGEP PKSGDRSGYS SPGSPGTPGS RSRTPSLPTP P TREPKKVA ...String: MAEPRQEFEV MEDHAGTYGL GDRKDQGGYT MHQDQEGDTD AGLKEEAGIG DTPSLEDEAA GHVTQARMVS KSKDGTGSDD KKAKGADGK TKIATPRGAA PPGQKGQANA TRIPAKTPPA PKTPPSSGEP PKSGDRSGYS SPGSPGTPGS RSRTPSLPTP P TREPKKVA VVRTPPKSPS SAKSRLQTAP VPMPDLKNVK SKIGSTENLK HQPGGGKVQI INKKLDLSNV QSKCGSKDNI KH VPGGGSV QIVYKPVDLS KVTSKCGSLG NIHHKPGGGQ VEVKSEKLDF KDRVQSKIGS LDNITHVPGG GNKKIETHKL TFR ENAKAK TDHGAEIVYK SPVVSGDTSP RHLSNVSSTG SIDMVDSPQL ATLADEVSAS LAKQGL UniProtKB: Microtubule-associated protein tau |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







































































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)