+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Plunge-frozen (control) map of beta-galactosidase | |||||||||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||||||||
 Keywords Keywords | Lactase / Beta-galactosidase / cryo-EM / native MS / HYDROLASE | |||||||||||||||||||||||||||||||||
| Biological species |   | |||||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.3 Å | |||||||||||||||||||||||||||||||||
 Authors Authors | Esser TK / Boehning J / Bharat TAM / Rauschenbach S | |||||||||||||||||||||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  France, France,  United States, European Union, 10 items United States, European Union, 10 items
| |||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Cryo-EM of soft-landed β-galactosidase: Gas-phase and native structures are remarkably similar. Authors: Tim K Esser / Jan Böhning / Alpcan Önür / Dinesh K Chinthapalli / Lukas Eriksson / Marko Grabarics / Paul Fremdling / Albert Konijnenberg / Alexander Makarov / Aurelien Botman / Christine ...Authors: Tim K Esser / Jan Böhning / Alpcan Önür / Dinesh K Chinthapalli / Lukas Eriksson / Marko Grabarics / Paul Fremdling / Albert Konijnenberg / Alexander Makarov / Aurelien Botman / Christine Peter / Justin L P Benesch / Carol V Robinson / Joseph Gault / Lindsay Baker / Tanmay A M Bharat / Stephan Rauschenbach /     Abstract: Native mass spectrometry (MS) has become widely accepted in structural biology, providing information on stoichiometry, interactions, homogeneity, and shape of protein complexes. Yet, the fundamental ...Native mass spectrometry (MS) has become widely accepted in structural biology, providing information on stoichiometry, interactions, homogeneity, and shape of protein complexes. Yet, the fundamental assumption that proteins inside the mass spectrometer retain a structure faithful to native proteins in solution remains a matter of intense debate. Here, we reveal the gas-phase structure of β-galactosidase using single-particle cryo-electron microscopy (cryo-EM) down to 2.6-Å resolution, enabled by soft landing of mass-selected protein complexes onto cold transmission electron microscopy (TEM) grids followed by in situ ice coating. We find that large parts of the secondary and tertiary structure are retained from the solution. Dehydration-driven subunit reorientation leads to consistent compaction in the gas phase. By providing a direct link between high-resolution imaging and the capability to handle and select protein complexes that behave problematically in conventional sample preparation, the approach has the potential to expand the scope of both native mass spectrometry and cryo-EM. | |||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18245.map.gz emd_18245.map.gz | 259.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18245-v30.xml emd-18245-v30.xml emd-18245.xml emd-18245.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18245_fsc.xml emd_18245_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_18245.png emd_18245.png | 95.9 KB | ||
| Filedesc metadata |  emd-18245.cif.gz emd-18245.cif.gz | 6.5 KB | ||
| Others |  emd_18245_half_map_1.map.gz emd_18245_half_map_1.map.gz emd_18245_half_map_2.map.gz emd_18245_half_map_2.map.gz | 254.5 MB 254.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18245 http://ftp.pdbj.org/pub/emdb/structures/EMD-18245 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18245 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18245 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18245.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18245.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
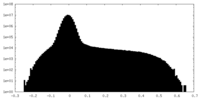
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_18245_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
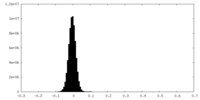
| Density Histograms |
-Half map: #2
| File | emd_18245_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
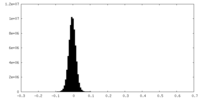
| Density Histograms |
- Sample components
Sample components
-Entire : Beta-galactosidase from E. coli; tetrameric complex
| Entire | Name: Beta-galactosidase from E. coli; tetrameric complex |
|---|---|
| Components |
|
-Supramolecule #1: Beta-galactosidase from E. coli; tetrameric complex
| Supramolecule | Name: Beta-galactosidase from E. coli; tetrameric complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Sigma Aldrich Product Number G3153 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 465 KDa |
-Macromolecule #1: Beta-galactosidase
| Macromolecule | Name: Beta-galactosidase / type: other / ID: 1 / Classification: other |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MTMITDSLAV VLQRRDWENP GVTQLNRLAA HPPFASWRNS EEARTDRPSQ QLRSLNGEWR FAWFPAPEA VPESWLECDL PEADTVVVPS NWQMHGYDAP IYTNVTYPIT VNPPFVPTEN P TGCYSLTF NVDESWLQEG QTRIIFDGVN SAFHLWCNGR WVGYGQDSRL ...String: MTMITDSLAV VLQRRDWENP GVTQLNRLAA HPPFASWRNS EEARTDRPSQ QLRSLNGEWR FAWFPAPEA VPESWLECDL PEADTVVVPS NWQMHGYDAP IYTNVTYPIT VNPPFVPTEN P TGCYSLTF NVDESWLQEG QTRIIFDGVN SAFHLWCNGR WVGYGQDSRL PSEFDLSAFL RA GENRLAV MVLRWSDGSY LEDQDMWRMS GIFRDVSLLH KPTTQISDFH VATRFNDDFS RAV LEAEVQ MCGELRDYLR VTVSLWQGET QVASGTAPFG GEIIDERGGY ADRVTLRLNV ENPK LWSAE IPNLYRAVVE LHTADGTLIE AEACDVGFRV VRIENGLLLL NGKPLLIRGV NRHEH HPLH GQVMDEQTMV QDILLMKQNN FNAVRCSHYP NHPLWYTLCD RYGLYVVDEA NIETHG MVP MNRLTDDPRW LPAMSERVTR MVQRDRNHPS VIIWSLGNES GHGANHDALY RWIKSVD PS RPVQYEGGGA DTTATDIICP MYARVDEDQP FPAVPKWSIK KWLSLPGETR PLILCEYA H AMGNSLGGFA KYWQAFRQYP RLQGGFVWDW VDQSLIKYDE NGNPWSAYGG DFGDTPNDR QFCMNGLVFA DRTPHPALTE AKHQQQFFQF RLSGQTIEVT SEYLFRHSDN ELLHWMVALD GKPLASGEV PLDVAPQGKQ LIELPELPQP ESAGQLWLTV RVVQPNATAW SEAGHISAWQ Q WRLAENLS VTLPAASHAI PHLTTSEMDF CIELGNKRWQ FNRQSGFLSQ MWIGDKKQLL TP LRDQFTR APLDNDIGVS EATRIDPNAW VERWKAAGHY QAEAALLQCT ADTLADAVLI TTA HAWQHQ GKTLFISRKT YRIDGSGQMA ITVDVEVASD TPHPARIGLN CQLAQVAERV NWLG LGPQE NYPDRLTAAC FDRWDLPLSD MYTPYVFPSE NGLRCGTREL NYGPHQWRGD FQFNI SRYS QQQLMETSHR HLLHAEEGTW LNIDGFHMGI GGDDSWSPSV SAEFQLSAGR YHYQLV WCQ K |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.9 / Details: 25 mM Tris, 50 mM NaCl, 10 mM EDTA, 2 mM MgCl2 |
|---|---|
| Grid | Model: Quantifoil / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Details: Plunge-frozen. |
| Details | 2.5 uM protein concentration |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 4100 / Average electron dose: 34.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | The PDB model 6CVM is an excellent fit for this map. No refinement was performed. |
| Refinement | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)