[English] 日本語
 Yorodumi
Yorodumi- EMDB-18216: Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylate... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer | |||||||||
 Map data Map data | DeepEMhancer map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cullin-RING RBR E3 Ligase / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcullin-RING-type E3 NEDD8 transferase / NEDD8 transferase activity / cullin-RING ubiquitin ligase complex / cellular response to chemical stress / Cul7-RING ubiquitin ligase complex / ubiquitin-dependent protein catabolic process via the C-end degron rule pathway / Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling / positive regulation of protein autoubiquitination / RNA polymerase II transcription initiation surveillance / protein neddylation ...cullin-RING-type E3 NEDD8 transferase / NEDD8 transferase activity / cullin-RING ubiquitin ligase complex / cellular response to chemical stress / Cul7-RING ubiquitin ligase complex / ubiquitin-dependent protein catabolic process via the C-end degron rule pathway / Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling / positive regulation of protein autoubiquitination / RNA polymerase II transcription initiation surveillance / protein neddylation / NEDD8 ligase activity / VCB complex / negative regulation of response to oxidative stress / Cul5-RING ubiquitin ligase complex / SCF ubiquitin ligase complex / Cul2-RING ubiquitin ligase complex / negative regulation of type I interferon production / ubiquitin-ubiquitin ligase activity / Cul4A-RING E3 ubiquitin ligase complex / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / Cul4-RING E3 ubiquitin ligase complex / Cul3-RING ubiquitin ligase complex / regulation of mitotic nuclear division / Cul4B-RING E3 ubiquitin ligase complex / negative regulation of mitophagy / Prolactin receptor signaling / TGF-beta receptor signaling activates SMADs / cullin family protein binding / regulation of proteolysis / regulation of postsynapse assembly / anatomical structure morphogenesis / protein monoubiquitination / protein K48-linked ubiquitination / Nuclear events stimulated by ALK signaling in cancer / Regulation of BACH1 activity / transcription-coupled nucleotide-excision repair / regulation of cellular response to insulin stimulus / positive regulation of TORC1 signaling / post-translational protein modification / negative regulation of insulin receptor signaling pathway / T cell activation / Degradation of DVL / Iron uptake and transport / Recognition of DNA damage by PCNA-containing replication complex / Degradation of GLI1 by the proteasome / GSK3B and BTRC:CUL1-mediated-degradation of NFE2L2 / Negative regulation of NOTCH4 signaling / Vif-mediated degradation of APOBEC3G / Hedgehog 'on' state / Degradation of GLI2 by the proteasome / GLI3 is processed to GLI3R by the proteasome / DNA Damage Recognition in GG-NER / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / cellular response to amino acid stimulus / Degradation of beta-catenin by the destruction complex / Evasion by RSV of host interferon responses / modification-dependent protein catabolic process / Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha / NOTCH1 Intracellular Domain Regulates Transcription / Dual Incision in GG-NER / Transcription-Coupled Nucleotide Excision Repair (TC-NER) / Formation of TC-NER Pre-Incision Complex / Constitutive Signaling by NOTCH1 PEST Domain Mutants / Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants / negative regulation of canonical Wnt signaling pathway / Formation of Incision Complex in GG-NER / protein tag activity / Regulation of expression of SLITs and ROBOs / RING-type E3 ubiquitin transferase / Interleukin-1 signaling / Orc1 removal from chromatin / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / protein modification process / Regulation of RAS by GAPs / microtubule cytoskeleton organization / Regulation of RUNX2 expression and activity / KEAP1-NFE2L2 pathway / UCH proteinases / protein polyubiquitination / positive regulation of protein catabolic process / Cargo recognition for clathrin-mediated endocytosis / Antigen processing: Ubiquitination & Proteasome degradation / cellular response to UV / ubiquitin protein ligase activity / intracellular protein localization / positive regulation of proteasomal ubiquitin-dependent protein catabolic process / Neddylation / MAPK cascade / cellular response to oxidative stress / ubiquitin-dependent protein catabolic process / spermatogenesis / molecular adaptor activity / proteasome-mediated ubiquitin-dependent protein catabolic process / Potential therapeutics for SARS / RNA polymerase II-specific DNA-binding transcription factor binding / positive regulation of canonical NF-kappaB signal transduction / postsynapse / protein ubiquitination / ubiquitin protein ligase binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Hopf LVM / Horn-Ghetko D / Schulman BA | |||||||||
| Funding support | European Union,  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nat.Struct.Mol.Biol. / Year: 2024 Journal: Nat.Struct.Mol.Biol. / Year: 2024Title: Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex. Authors: Horn-Ghetko D / Hopf LVM / Tripathi-Giesgen I / Du J / Kostrhon S / Vu DT / Beier V / Steigenberger B / Prabu JR / Stier L / Bruss EM / Mann M / Xiong Y / Schulman BA | |||||||||
| History |
|
- Structure visualization
Structure visualization
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18216.map.gz emd_18216.map.gz | 433.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18216-v30.xml emd-18216-v30.xml emd-18216.xml emd-18216.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18216_fsc.xml emd_18216_fsc.xml | 17.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_18216.png emd_18216.png | 133.5 KB | ||
| Masks |  emd_18216_msk_1.map emd_18216_msk_1.map | 488.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18216.cif.gz emd-18216.cif.gz | 7.8 KB | ||
| Others |  emd_18216_additional_1.map.gz emd_18216_additional_1.map.gz emd_18216_half_map_1.map.gz emd_18216_half_map_1.map.gz emd_18216_half_map_2.map.gz emd_18216_half_map_2.map.gz | 31 MB 391.2 MB 390.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18216 http://ftp.pdbj.org/pub/emdb/structures/EMD-18216 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18216 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18216 | HTTPS FTP |
-Validation report
| Summary document |  emd_18216_validation.pdf.gz emd_18216_validation.pdf.gz | 777.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_18216_full_validation.pdf.gz emd_18216_full_validation.pdf.gz | 777.4 KB | Display | |
| Data in XML |  emd_18216_validation.xml.gz emd_18216_validation.xml.gz | 25.7 KB | Display | |
| Data in CIF |  emd_18216_validation.cif.gz emd_18216_validation.cif.gz | 34.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18216 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18216 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18216 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-18216 | HTTPS FTP |
-Related structure data
| Related structure data | 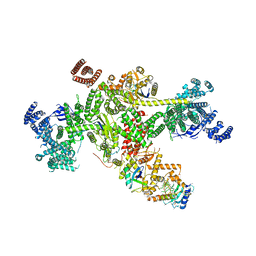 8q7hMC 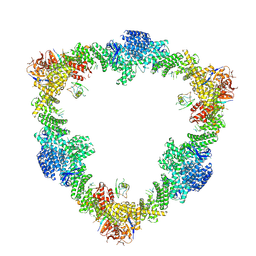 8q7eC 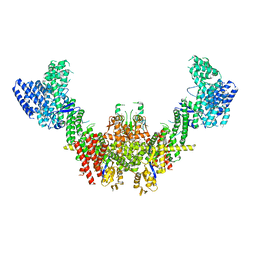 8rhzC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18216.map.gz / Format: CCP4 / Size: 488.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18216.map.gz / Format: CCP4 / Size: 488.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8512 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylate...
| Entire | Name: Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer |
|---|---|
| Components |
|
-Supramolecule #1: Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylate...
| Supramolecule | Name: Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 / Details: Structure of dimeric subunit |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Cullin-9
| Macromolecule | Name: Cullin-9 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 281.686062 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPMVGERHAG DLMVPLGPRL QAYPEELIRQ RPGHDGHPEY LIRWSVLKCG EVGKVGVEEG KAEHILMWLS APEVYANCPG LLGERALSK GLQHEPAGVS GSFPRDPGGL DEVAMGEMEA DVQALVRRAA RQLAESGTPS LTAAVLHTIH VLSAYASIGP L TGVFRETG ...String: GPMVGERHAG DLMVPLGPRL QAYPEELIRQ RPGHDGHPEY LIRWSVLKCG EVGKVGVEEG KAEHILMWLS APEVYANCPG LLGERALSK GLQHEPAGVS GSFPRDPGGL DEVAMGEMEA DVQALVRRAA RQLAESGTPS LTAAVLHTIH VLSAYASIGP L TGVFRETG ALDLLMHMLC NPEPQIRRSA GKMLQALAAH DAGSRAHVLL SLSQQDGIEQ HMDFDSRYTL LELFAETTSS EE HCMAFEG IHLPQIPGKL LFSLVKRYLC VTSLLDQLNS SPELGAGDQS SPCATREKSR GQRELEFSMA VGNLISELVR SMG WARNLS EQGMSPPRPT RSIFQPYISG PSLLLPTIVT TPRRQGWVFR QRSEFSSRSG YGEYVQQTLQ PGMRVRMLDD YEEI SAGDE GEFRQSNNGI PPVQVFWQST GRTYWVHWHM LEILGPEEAT EDKASAAVEK GAGATVLGTA FPSWDWNPMD GLYPL PYLQ PEPQKNERVG YLTQAEWWEL LFFIKKLDLC EQQPIFQNLW KNLDETLGEK ALGEISVSVE MAESLLQVLS SRFEGS TLN DLLNSQIYTK YGLLSNEPSS SSTSRNHSCT PDPEEESKSE ASFSEEETES LKAKAEAPKT EAEPTKTRTE TPMAQSD SQ LFNQLLVTEG MTLPTEMKEA ASEMARALRG PGPRSSLDQH VAAVVATVQI SSLDTNLQLS GLSALSQAVE EVTERDHP L VRPDRSLREK LVKMLVELLT NQVGEKMVVV QALRLLYLLM TKHEWRPLFA REGGIYAVLV CMQEYKTSVL VQQAGLAAL KMLAVASSSE IPTFVTGRDS IHSLFDAQMT REIFASIDSA TRPGSESLLL TVPAAVILML NTEGCSSAAR NGLLLLNLLL CNHHTLGDQ IITQELRDTL FRHSGIAPRT EPMPTTRTIL MMLLNRYSEP PGSPERAALE TPIIQGQDGS PELLIRSLVG G PSAELLLD LERVLCREGS PGGAVRPLLK RLQQETQPFL LLLRTLDAPG PNKTLLLSVL RVITRLLDFP EAMVLPWHEV LE PCLNCLS GPSSDSEIVQ ELTCFLHRLA SMHKDYAVVL CCLGAKEILS KVLDKHSAQL LLGCELRDLV TECEKYAQLY SNL TSSILA GCIQMVLGQI EDHRRTHQPI NIPFFDVFLR HLCQGSSVEV KEDKCWEKVE VSSNPHRASK LTDHNPKTYW ESNG STGSH YITLHMHRGV LVRQLTLLVA SEDSSYMPAR VVVFGGDSTS CIGTELNTVN VMPSASRVIL LENLNRFWPI IQIRI KRCQ QGGIDTRVRG VEVLGPKPTF WPLFREQLCR RTCLFYTIRA QAWSRDIAED HRRLLQLCPR LNRVLRHEQN FADRFL PDD EAAQALGKTC WEALVSPLVQ NITSPDAEGV SALGWLLDQY LEQRETSRNP LSRAASFASR VRRLCHLLVH VEPPPGP SP EPSTRPFSKN SKGRDRSPAP SPVLPSSSLR NITQCWLSVV QEQVSRFLAA AWRAPDFVPR YCKLYEHLQR AGSELFGP R AAFMLALRSG FSGALLQQSF LTAAHMSEQF ARYIDQQIQG GLIGGAPGVE MLGQLQRHLE PIMVLSGLEL ATTFEHFYQ HYMADRLLSF GSSWLEGAVL EQIGLCFPNR LPQLMLQSLS TSEELQRQFH LFQLQRLDKL FLEQEDEEEK RLEEEEEEEE EEEAEKELF IEDPSPAISI LVLSPRCWPV SPLCYLYHPR KCLPTEFCDA LDRFSSFYSQ SQNHPVLDMG PHRRLQWTWL G RAELQFGK QILHVSTVQM WLLLKFNQTE EVSVETLLKD SDLSPELLLQ ALVPLTSGNG PLTLHEGQDF PHGGVLRLHE PG PQRSGEA LWLIPPQAYL NVEKDEGRTL EQKRNLLSCL LVRILKAHGE KGLHIDQLVC LVLEAWQKGP NPPGTLGHTV AGG VACTST DVLSCILHLL GQGYVKRRDD RPQILMYAAP EPMGPCRGQA DVPFCGSQSE TSKPSPEAVA TLASLQLPAG RTMS PQEVE GLMKQTVRQV QETLNLEPDV AQHLLAHSHW GAEQLLQSYS EDPEPLLLAA GLCVHQAQAV PVRPDHCPVC VSPLG CDDD LPSLCCMHYC CKSCWNEYLT TRIEQNLVLN CTCPIADCPA QPTGAFIRAI VSSPEVISKY EKALLRGYVE SCSNLT WCT NPQGCDRILC RQGLGCGTTC SKCGWASCFN CSFPEAHYPA SCGHMSQWVD DGGYYDGMSV EAQSKHLAKL ISKRCPS CQ APIEKNEGCL HMTCAKCNHG FCWRCLKSWK PNHKDYYNCS AMVSKAARQE KRFQDYNERC TFHHQAREFA VNLRNRVS A IHEVPPPRSF TFLNDACQGL EQARKVLAYA CVYSFYSQDA EYMDVVEQQT ENLELHTNAL QILLEETLLR CRDLASSLR LLRADCLSTG MELLRRIQER LLAILQHSAQ DFRVGLQSPS VEAWEAKGPN MPGSQPQASS GPEAEEEEED DEDDVPEWQQ DEFDEELDN DSFSYDESEN LDQETFFFGD EEEDEDEAYD UniProtKB: Cullin-9 |
-Macromolecule #2: E3 ubiquitin-protein ligase RBX1
| Macromolecule | Name: E3 ubiquitin-protein ligase RBX1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.289977 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAAMDVDTP SGTNSGAGKK RFEVKKWNAV ALWAWDIVVD NCAICRNHIM DLCIECQANQ ASATSEECTV AWGVCNHAFH FHCISRWLK TRQVCPLDNR EWEFQKYGH UniProtKB: E3 ubiquitin-protein ligase RBX1 |
-Macromolecule #3: NEDD8
| Macromolecule | Name: NEDD8 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.086562 KDa |
| Sequence | String: MLIKVKTLTG KEIEIDIEPT DKVERIKERV EEKEGIPPQQ QRLIYSGKQM NDEKTAADYK ILGGSVLHLV LALRGGGGLR Q UniProtKB: Ubiquitin-like protein NEDD8 |
-Macromolecule #4: Unknown
| Macromolecule | Name: Unknown / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.868854 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 10 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 71928 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
















