+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | X. laevis CMG dimer bound to dimeric DONSON - MCM ATPase | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | DNA replication initiation / Xenopus egg extract / primordial dwarfism / replicative helicase / genome stability / REPLICATION | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcell cycle DNA replication / single-stranded 3'-5' DNA helicase activity / nuclear DNA replication / premeiotic DNA replication / mitotic DNA replication / CMG complex / MCM complex / mitotic DNA replication checkpoint signaling / double-strand break repair via break-induced replication / mitotic DNA replication initiation ...cell cycle DNA replication / single-stranded 3'-5' DNA helicase activity / nuclear DNA replication / premeiotic DNA replication / mitotic DNA replication / CMG complex / MCM complex / mitotic DNA replication checkpoint signaling / double-strand break repair via break-induced replication / mitotic DNA replication initiation / replisome / single-stranded DNA helicase activity / regulation of DNA-templated DNA replication initiation / DNA strand elongation involved in DNA replication / 3'-5' DNA helicase activity / mitotic G2 DNA damage checkpoint signaling / replication fork processing / DNA replication origin binding / DNA replication initiation / DNA damage checkpoint signaling / replication fork / single-stranded DNA binding / DNA helicase / DNA replication / cell division / chromatin binding / chromatin / ATP hydrolysis activity / zinc ion binding / ATP binding / nucleus Similarity search - Function | ||||||||||||||||||
| Biological species | |||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.53 Å | ||||||||||||||||||
 Authors Authors | Cvetkovic MA / Costa A | ||||||||||||||||||
| Funding support |  United Kingdom, European Union, 5 items United Kingdom, European Union, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: The structural mechanism of dimeric DONSON in replicative helicase activation. Authors: Milos A Cvetkovic / Paolo Passaretti / Agata Butryn / Alicja Reynolds-Winczura / Georgia Kingsley / Aggeliki Skagia / Cyntia Fernandez-Cuesta / Divyasree Poovathumkadavil / Roger George / ...Authors: Milos A Cvetkovic / Paolo Passaretti / Agata Butryn / Alicja Reynolds-Winczura / Georgia Kingsley / Aggeliki Skagia / Cyntia Fernandez-Cuesta / Divyasree Poovathumkadavil / Roger George / Anoop S Chauhan / Satpal S Jhujh / Grant S Stewart / Agnieszka Gambus / Alessandro Costa /  Abstract: The MCM motor of the replicative helicase is loaded onto origin DNA as an inactive double hexamer before replication initiation. Recruitment of activators GINS and Cdc45 upon S-phase transition ...The MCM motor of the replicative helicase is loaded onto origin DNA as an inactive double hexamer before replication initiation. Recruitment of activators GINS and Cdc45 upon S-phase transition promotes the assembly of two active CMG helicases. Although work with yeast established the mechanism for origin activation, how CMG is formed in higher eukaryotes is poorly understood. Metazoan Downstream neighbor of Son (DONSON) has recently been shown to deliver GINS to MCM during CMG assembly. What impact this has on the MCM double hexamer is unknown. Here, we used cryoelectron microscopy (cryo-EM) on proteins isolated from replicating Xenopus egg extracts to identify a double CMG complex bridged by a DONSON dimer. We find that tethering elements mediating complex formation are essential for replication. DONSON reconfigures the MCM motors in the double CMG, and primordial dwarfism patients' mutations disrupting DONSON dimerization affect GINS and MCM engagement in human cells and DNA synthesis in Xenopus egg extracts. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18192.map.gz emd_18192.map.gz | 567.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18192-v30.xml emd-18192-v30.xml emd-18192.xml emd-18192.xml | 25.4 KB 25.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18192_fsc.xml emd_18192_fsc.xml | 17.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_18192.png emd_18192.png | 112 KB | ||
| Filedesc metadata |  emd-18192.cif.gz emd-18192.cif.gz | 9.1 KB | ||
| Others |  emd_18192_half_map_1.map.gz emd_18192_half_map_1.map.gz emd_18192_half_map_2.map.gz emd_18192_half_map_2.map.gz | 557.5 MB 557.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18192 http://ftp.pdbj.org/pub/emdb/structures/EMD-18192 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18192 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18192 | HTTPS FTP |
-Related structure data
| Related structure data |  8q6pMC  8q6oC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18192.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18192.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_18192_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
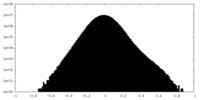
| Density Histograms |
-Half map: #1
| File | emd_18192_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
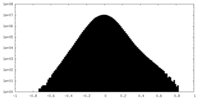
| Density Histograms |
- Sample components
Sample components
-Entire : X. laevis CMG dimer bound to dimeric DONSON
| Entire | Name: X. laevis CMG dimer bound to dimeric DONSON |
|---|---|
| Components |
|
-Supramolecule #1: X. laevis CMG dimer bound to dimeric DONSON
| Supramolecule | Name: X. laevis CMG dimer bound to dimeric DONSON / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 1.54 MDa |
-Macromolecule #1: DNA replication licensing factor mcm2
| Macromolecule | Name: DNA replication licensing factor mcm2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 100.397648 KDa |
| Sequence | String: MADSSESFNI ATSPRAGSRR DALTSSPGRD LPPFEDESEG MFGDGVVPEE EEDGEELIGD AMERDYRPIS ELDRYEVEGL DDEEDVEDL TASQREAAEQ SMRMRDREMG RELGRMRRGL LYDSDEEEED RPARKRRMAE RAAEGAPEED EEMIESIENL E DMKGHTVR ...String: MADSSESFNI ATSPRAGSRR DALTSSPGRD LPPFEDESEG MFGDGVVPEE EEDGEELIGD AMERDYRPIS ELDRYEVEGL DDEEDVEDL TASQREAAEQ SMRMRDREMG RELGRMRRGL LYDSDEEEED RPARKRRMAE RAAEGAPEED EEMIESIENL E DMKGHTVR EWVSMAATRL EIYHRFKNFL RTHVDEHGHN VFKEKISDMC KENKESLPVN YEDLAAREHV LAYFLPEAPA EM LKIFDEA AKEVVLVMYP KYDRIAREIH VRISHLPLVE ELRSLRQLHL NQLIRTSGVV TCCTGVLPQL SMVKYNCNKC NFI LGPFFQ SQNQEVRPGS CPECQSFGPF EINMEETVYQ NYQRITIQES PGKVAAGRLP RSKDAILLAD LVDSCKPGDE IELT GIYHN NYDGSLNTAN GFPVFATVIL ANHITKKDDK VAVGELTDED VKAIVALSKD ERIGERIFAS IAPSIYGHED IKRGL ALAL FGGEAKNPGG KHKVRGDINV LLCGDPGTAK SQFLKYVEKV ASRAVFTTGQ GASAVGLTAY VQRHPVTKEW TLEAGA LVL ADRGVCLIDE FDKMNDQDRT SIHEAMEQQS ISISKAGIVT SLQARCTVIA ASNPIGGRYD PSLTFSENVD LTEPIVS RF DILCVVRDTV DPVQDEMLAR FVVSSHIKHH PSSKDIANGD AAEFALPNTF GVEALPQEVL KKYIMYAKEK IRPKLNQM D QDKVAKMYSD LRKESMATGS IPITVRHIES MIRMAEAHAR MHLRDYVVED DVNMAIRVML ESFIDTQKFS VMRSMRKTF ARYLAFRRDN NELLLFVLKQ LIAEQVTYQR NRYGAQQDTI EVPEKDLVDK ARQINIHNLS AFYDSDLFKM NKFTHDVKKK LIIQQF UniProtKB: DNA replication licensing factor mcm2 |
-Macromolecule #2: Maternal DNA replication licensing factor mcm3
| Macromolecule | Name: Maternal DNA replication licensing factor mcm3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 90.5365 KDa |
| Sequence | String: MDYGGGFEDH ELREAQREYL DFLDDDQDQG LYHGKVRDMI GSNEHRLIVN LNDVRRKNDK RANLMLNDAF AETIAFQRAL KDLVASIDA TYAKQFEEFS VGFEGSFGSK HVSPRTLTAS LLGSLVCVEG IVTKCSLVRP KVMRSVHYCP ATKKTLERKY S DLTSLEAF ...String: MDYGGGFEDH ELREAQREYL DFLDDDQDQG LYHGKVRDMI GSNEHRLIVN LNDVRRKNDK RANLMLNDAF AETIAFQRAL KDLVASIDA TYAKQFEEFS VGFEGSFGSK HVSPRTLTAS LLGSLVCVEG IVTKCSLVRP KVMRSVHYCP ATKKTLERKY S DLTSLEAF PSSSIYPTKD EENNPLETEY GLSTYKDHQT LSIQEMPEKA PAGQLPRSVD IIADDDLVDK CKPGDRVQIV GI YRCLPSK QGGFTSGTFR TILLANNIKL MSKEIAPTFS ADDVAKIKKF CKAHSKDIFE HLSKSLAPSI HGHEYIKKAI LCM LLGGNE KVLENGTRIR GDINVLLIGD PSVAKSQLLR YVLHTAPRAI PTTGRGSSGV GLTAAVTTDQ ETGERRLEAG AMVL ADRGV VCIDEFDKMS DMDRTAIHEV MEQGRVTIAK AGIQARLNAR CSVLAAANPV YGRYDQYRTP MENIGLQDSL LSRFD LLFI VLDKMDADND QEIADHVLRM HRYRTPGEQD GYALPLGCSV EIFATDDPNA SDVTDQELQI YEKHDNLLHG PRKNKS KIV SMQFIRKYIH VAKLIKPVLT SEAADYISQE YAKIRNHDQI NNDSARTMPV TARALETMIR LSTAHAKVRM SKTIERQ DA ETALELVQFA YFKKVLAKEK KKTDKDLHDE NLSQDTLSQE SVRKSSRRAG KIADSQDDSM DPYSFSEQDS SLNENLSQ S LRPQRKKAES QDGKRSLSQN RTKEFKAALL KAFKSSRSQS VAVSQLLELI NKGNPEPFER SEVKEALDNM QNDNQVMVS EDVVFLI UniProtKB: Maternal DNA replication licensing factor mcm3 |
-Macromolecule #3: DNA replication licensing factor mcm4-B
| Macromolecule | Name: DNA replication licensing factor mcm4-B / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 97.256305 KDa |
| Sequence | String: MSSPTSTPSR RRNKRGRGSN PPTPHGEEVQ SPPSQRRRTE DSTSIGELLP MPTSPSGDVQ SPSGQELLFS SPAPSRHSAH QSELDLSSP LTYGTPSSRV EGTPRSGIRG TPARQRPDLG SARKVKQVDL HSDQPAAEEL VTSEQSLGQK LVIWGTDVNV A TCKEKFQR ...String: MSSPTSTPSR RRNKRGRGSN PPTPHGEEVQ SPPSQRRRTE DSTSIGELLP MPTSPSGDVQ SPSGQELLFS SPAPSRHSAH QSELDLSSP LTYGTPSSRV EGTPRSGIRG TPARQRPDLG SARKVKQVDL HSDQPAAEEL VTSEQSLGQK LVIWGTDVNV A TCKEKFQR FVQRFIDPSA KEEDNVGLDL NEPIYMQRLE EINVVGDPFL NIDCDHLRNF DQDLYRQLVC YPQEVIPTFD MA ANEIFFE RYPDSILEHQ IQVRPYNALK TRNMRSLNPE DIDQLITISG MVIRTSQIIP EMQEAFFKCQ VCAFTTRVEI DRG RIAEPS VCKHCNTTHS MALIHNRSMF SDKQMIKLQE SPEDMPAGQT PHTTILYGHN DLVDKVQPGD RVNVTGIYRA VPIR VNPRV RNVKSVYKTH IDVIHYRKTD SKRLHGIDED TEQKLFTEER VAMLKELAAK PDIYERLAAA LAPSIYEHED IKKGI LLQL FGGTRKDFSH TGRGKFRAEV NILLCGDPGT SKSQLLQYVF NLVPRGQYTS GKGSSAVGLT AYVMKDPETR QLVLQT GAL VLSDNGICCI DEFDKMNEST RSVLHEVMEQ QTLSIAKAGI ICQLNARTSV LAAANPVESQ WNPKKTTIEN IQLPHTL LS RFDLIFLMLD PQDEAYDRRL AHHLVALYYQ SEEQMKEEHL DMAVLKDYIA YARTYVNPRL SEEASQALIE AYVSMRKI G SGRGMVSAYP RQLESLIRLS EAHAKVRFSN KVETIDVEEA KRLHREALKQ SATDPRTGIV DISILTTGMS ATARKRKEE LAQVLKKLIQ SKGKTPALKY QQLFEDLRGQ SDAAITKDMF DEALHALADD DYLTVTGKTV RLL UniProtKB: DNA replication licensing factor mcm4-B |
-Macromolecule #4: DNA replication licensing factor mcm5-A
| Macromolecule | Name: DNA replication licensing factor mcm5-A / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 82.556414 KDa |
| Sequence | String: MSGFDDLGVY YSDSFGGEQQ VGDDGQAKKS QLKKRFREFL RQYRIGTDRT GFTFKYRDEL KRHYNLGEYW IEVEMEDLAS FDEDLADYL YKQPTEHLQL LEEAAQEVAD EVTRPRPAGE ETIQEIQVML RSDANPANIR SLKSEQMSHL VKIPGIIIAA T AVRAKATK ...String: MSGFDDLGVY YSDSFGGEQQ VGDDGQAKKS QLKKRFREFL RQYRIGTDRT GFTFKYRDEL KRHYNLGEYW IEVEMEDLAS FDEDLADYL YKQPTEHLQL LEEAAQEVAD EVTRPRPAGE ETIQEIQVML RSDANPANIR SLKSEQMSHL VKIPGIIIAA T AVRAKATK ISIQCRSCRN TIGNIAVRPG LEGYAMPRKC NTEQAGRPNC PLDPYFIIPD KCKCVDFQTL KLQESPDAVP HG ELPRHMQ LYCDRYLCDK VVPGNRVTIM GIYSIRKSGK TSTKGRDRVG VGIRSSYIRV VGIQVDTEGT GRSAAGAITP QEE EEFRRL AAKPDIYETV AKSIAPSIYG SSDIKKAIAC LLFGGSRKRL PDGLTRRGDV NLLMLGDPGT AKSQLLKFVE RCSP IGVYT SGKGSSAAGL TASVMRDPVS RNFIMEGGAM VLADGGVVCI DEFDKMREDD RVAIHEAMEQ QTISIAKAGI TTTLN SRCS VLAAANSVYG RWDDTKGEEN IDFMPTILSR FDMIFIVKDE HNEQRDMTLA KHVMNVHLSA RTQSSSVEGE VDLNTL KKY IAYCRAKCGP RLSAEAAEKL KNRYILMRSG AREHERETEK RSSIPITVRQ LEAIVRISES LGKMKLQPFA TETDVEE AL RLFQVSTLDA AMSGSLSGVE GFTTQEDQEM LSRIEKQMKK RFAIGSQVSE HSIIQDFLKQ KYPEHAIHKV LSLMMRRG E IQHRLQRKVL YRIK UniProtKB: DNA replication licensing factor mcm5-A |
-Macromolecule #5: Maternal DNA replication licensing factor mcm6
| Macromolecule | Name: Maternal DNA replication licensing factor mcm6 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 92.752969 KDa |
| Sequence | String: MELGGPAAAG DTDIAGQQLF KDELSDKCQK LFLEFLEECK GKDGSNLYVS AAEELIRPER NTLAVNFTDI EYYNQQLATT IQEEYYRVY PHLCRAVRSF ARQMGNIPAN KEFYIAFSDF PARQKIRELS SAKIGTLLRI SGQVVRTHPV HPELVSGTFL C MDCQSIVK ...String: MELGGPAAAG DTDIAGQQLF KDELSDKCQK LFLEFLEECK GKDGSNLYVS AAEELIRPER NTLAVNFTDI EYYNQQLATT IQEEYYRVY PHLCRAVRSF ARQMGNIPAN KEFYIAFSDF PARQKIRELS SAKIGTLLRI SGQVVRTHPV HPELVSGTFL C MDCQSIVK DVEQQFRYTQ PTICKNPVCA NRRRFTLDTN KSRFVDFQKV RIQETQAELP RGAIPRSVEI ILRAEAVESA MA GDRCDFT GTLIVVPDVS ALAAGDARME TGAKVTGGEG FNSEGVQGLK ALGVRDLSYR LAFLACYVGA TNPRFGGKDL REE DQTAES IKNQMTVQEW EKVFEMSQDK NLYHNLCTSL FPTIHGNDEI KRGVLLMLFG GVPKTTMEGT SLRGDINVCI VGDP STSKS QFLKHVEEFS PRAVYTSGKA SSAAGLTAAV VKDEESHEFV IEAGALMLAD NGVCCIDEFD KMDLKDQVAI HEAME QQTI SITKAGVKAT LNARTSILAA ANPVGGRYER SKSLKHNVNL SAPIMSRFDL FFILVDECNE VTDYAIARRI VDLHAR NEE SIERVYSIED IQRYLLFARQ FQPKITKEAE EFIVEQYRRL RQRDGSGVAK SSWRITVRQL ESLIRLSESM ARMHCSD EV QPKHVKEAFR LLSKSIIRVD TPDVSFDQGE DEKNIEGENN GNLNNGEEAM ETNQDEPINE KPSSNAGLKM SFAEYKQI S NLLVLYMQKM EETEEECHLT TTDLVNWYLK EMEAEIETET ELILKKRLIE KVIHRLIYYD HILIELNKSE LKTMDDTKE TGEDAAEDRI LVVNPNYMLE D UniProtKB: Maternal DNA replication licensing factor mcm6 |
-Macromolecule #6: DNA replication licensing factor mcm7-B
| Macromolecule | Name: DNA replication licensing factor mcm7-B / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 81.841539 KDa |
| Sequence | String: MPRDYQAEKE KCKTFLQEFY KDDEFGKKNF KYGVQLANIA HREQVALCID LDDLAEEDPE LVDAICENTR RYTNLFADAV QELLPQYKE REVVHKDALD VYIEHRLMME QRGRDPNEMR DPHNQYPPEL MRRFELYFKA PSSSKARVVR DVKADSIGKL V TVRGIVTR ...String: MPRDYQAEKE KCKTFLQEFY KDDEFGKKNF KYGVQLANIA HREQVALCID LDDLAEEDPE LVDAICENTR RYTNLFADAV QELLPQYKE REVVHKDALD VYIEHRLMME QRGRDPNEMR DPHNQYPPEL MRRFELYFKA PSSSKARVVR DVKADSIGKL V TVRGIVTR VTEVKPMMVV ATYTCDQCGA ETYQPIQSPT FMPLIMCPSR ECQTNRSGGR LYLQTRGSKF IKFQELKIQE HS DQVPVGN IPRCMSVYVR GENTRLAQPG DHVGITGVFL PMLRTGFRQV VQGLLSETYL ESHRLVKMNK TEDDELGTEE LSE EELRQI TEEDFYEKLA ASIAPEIYGH EDVKKALLLL LVGGVDHSPR GMKIRGNINV CLMGDPGVAK SQLLSYIDRL APRS QYTTG RGSSGVGLTA AVMKDPVTGE MTLEGGALVL ADQGVCCIDE FDKMMDSDRT AIHEVMEQQT ISIAKAGIMT TLNAR CSIL AAANPAYGRY NPKKTVEQNI QLPAALLSRF DLLWLIQDKP DRDNDLRLAQ HITYVHQHSK QPPSQFQPMD MKLMRR YIT MCKSKQPAIP ESLADYLTAA YVEMRKEART NKDMTFTSAR TLLSILRLST ALARLRLEDV VEKEDVNEAM RLTEMSK DS LQGDKGHASR TQRPADVIFS TIREMVPEKG ARSVKYSEAE QRCVSKGFTP AQFEAALEEY EELNVWLVNQ ARTKITFV UniProtKB: DNA replication licensing factor mcm7-B |
-Macromolecule #7: Protein downstream neighbor of son homolog
| Macromolecule | Name: Protein downstream neighbor of son homolog / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 63.864559 KDa |
| Sequence | String: MAELLPGYSP SFKKPSEILR LSRRRSRSEA SKTGLSPFSP GDVIKRVPGL RPFSPGPNKG AGVKRRNPFA SLENTVCSPV KRRAETVAE YGAASLEPRA LSAVRPSCLG QDSPEPPQFN SVEDVIWGDP LAADADPLVK TESPEKPAPA CEIPKGSVTF P ADWSLKTR ...String: MAELLPGYSP SFKKPSEILR LSRRRSRSEA SKTGLSPFSP GDVIKRVPGL RPFSPGPNKG AGVKRRNPFA SLENTVCSPV KRRAETVAE YGAASLEPRA LSAVRPSCLG QDSPEPPQFN SVEDVIWGDP LAADADPLVK TESPEKPAPA CEIPKGSVTF P ADWSLKTR LLFTSSHSFS WADHLKAQEE AQGLVMQCRA TAVNLPHSIQ EPKLSTDLRC AFQQSLVHWI HPSLPWVQLF PR IGVDRKM AGKNTPWSQD ESLQQVLMSE WALSFTSLYN LLKAKLCPYF YVCTYQFTVL FRAAGLAGSD VITAVMSPTT RGL REAMKN EGITFSQPLV EDDTGKKQKK PEAASQGDIN PEKENGTAEA DEASDESDED ESFSWLEEMG VEDKIKKPDS ISIK LRKEK NEVKLDHKPE SVVLVKGTNT FTLLNFLINC KSIVAAAGLQ AGLPPTLLSP VAFRGATMHA LKARSVNVKT RVNSG YKDQ FSLEITGPIM PHSLHSLTML LQSAQRGSFS AGLYTHEPTA VFNTPIHSQA VKEISADLQN CGLHPCTVEQ LTQVNE LGK LSLRHLEMTD YRYTWK UniProtKB: Protein downstream neighbor of son homolog |
-Macromolecule #8: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 8 / Number of copies: 5 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 33.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.4 µm / Nominal defocus min: 1.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)