[English] 日本語
 Yorodumi
Yorodumi- EMDB-18132: Bacterial transcription termination factor Rho G150D mutant bound... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacterial transcription termination factor Rho G150D mutant bound to ADP; C-terminal 8xHis-tag | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Rho / termination / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationATP-dependent activity, acting on RNA / DNA-templated transcription termination / helicase activity / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / ATP hydrolysis activity / RNA binding / ATP binding / identical protein binding / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Said N / Hilal T / Wahl MC | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: Transcription termination factor ρ polymerizes under stress. Authors: Bing Wang / Nelly Said / Tarek Hilal / Mark Finazzo / Markus C Wahl / Irina Artsimovitch /   Abstract: Bacterial RNA helicase ρ is a genome sentinel that terminates synthesis of damaged and junk RNAs that are not translated by the ribosome. Co-transcriptional RNA surveillance by ρ is essential for ...Bacterial RNA helicase ρ is a genome sentinel that terminates synthesis of damaged and junk RNAs that are not translated by the ribosome. Co-transcriptional RNA surveillance by ρ is essential for quality control of the transcriptome during optimal growth. However, it is unclear how bacteria protect their RNAs from overzealous ρ during dormancy or stress, conditions common in natural habitats. Here we used cryogenic electron microscopy, biochemical, and genetic approaches to show that residue substitutions, ADP, or ppGpp promote hyper-oligomerization of ρ. Our results demonstrate that nucleotides bound at subunit interfaces control ρ switching from active hexamers to inactive higher-order oligomers and extended filaments. Polymers formed upon exposure to antibiotics or ppGpp disassemble when stress is relieved, thereby directly linking termination activity to cellular physiology. Inactivation of ρ through hyper-oligomerization is a regulatory strategy shared by RNA polymerases, ribosomes, and metabolic enzymes across all life. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18132.map.gz emd_18132.map.gz | 52.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18132-v30.xml emd-18132-v30.xml emd-18132.xml emd-18132.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18132_fsc.xml emd_18132_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_18132.png emd_18132.png | 86.9 KB | ||
| Filedesc metadata |  emd-18132.cif.gz emd-18132.cif.gz | 5.9 KB | ||
| Others |  emd_18132_half_map_1.map.gz emd_18132_half_map_1.map.gz emd_18132_half_map_2.map.gz emd_18132_half_map_2.map.gz | 200.3 MB 200.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18132 http://ftp.pdbj.org/pub/emdb/structures/EMD-18132 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18132 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18132 | HTTPS FTP |
-Related structure data
| Related structure data |  8q3pMC  8q3nC  8q3oC  8q3qC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_18132.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18132.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
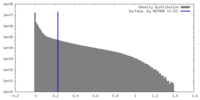
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_18132_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
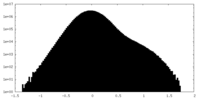
| Density Histograms |
-Half map: #1
| File | emd_18132_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
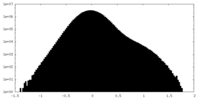
| Density Histograms |
- Sample components
Sample components
-Entire : Filament of Rho mutant G150D
| Entire | Name: Filament of Rho mutant G150D |
|---|---|
| Components |
|
-Supramolecule #1: Filament of Rho mutant G150D
| Supramolecule | Name: Filament of Rho mutant G150D / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Transcription termination factor Rho
| Macromolecule | Name: Transcription termination factor Rho / type: protein_or_peptide / ID: 1 / Number of copies: 18 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.616809 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHMNLTELK NTPVSELITL GENMGLENLA RMRKQDIIFA ILKQHAKSGE DIFGDGVLEI LQDGFGFLRS ADSSYLAGPD DIYVSPSQI RRFNLRTGDT ISGKIRPPKE GERYFALLKV NEVNFDKPEN ARNKILFENL TPLHANSRLR MERDNGSTED L TARVLDLA ...String: MGHMNLTELK NTPVSELITL GENMGLENLA RMRKQDIIFA ILKQHAKSGE DIFGDGVLEI LQDGFGFLRS ADSSYLAGPD DIYVSPSQI RRFNLRTGDT ISGKIRPPKE GERYFALLKV NEVNFDKPEN ARNKILFENL TPLHANSRLR MERDNGSTED L TARVLDLA SPIGRGQRGL IVAPPKAGKT MLLQNIAQSI AYNHPDCVLM VLLIDERPEE VTEMQRLVKG EVVASTFDEP AS RHVQVAE MVIEKAKRLV EHKKDVIILL DSITRLARAY NTVVPASGKV LTGGVDANAL HRPKRFFGAA RNVEEGGSLT IIA TALIDT GSKMDEVIYE EFKGTGNMEL HLSRKIAEKR VFPAIDYNRS GTRKEELLTT QEELQKMWIL RKIIHPMGEI DAME FLINK LAMTKTNDDF FEMMKRSGHH HHHHHH UniProtKB: Transcription termination factor Rho |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 18 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 18 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.4 mg/mL |
|---|---|
| Buffer | pH: 7.6 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2511 / Average exposure time: 40.57 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.9 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 96.4 |
|---|---|
| Output model |  PDB-8q3p: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)