+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hepatitis B core protein with bound Geraniol | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | geraniol Hepatitis B core protein hyfrophobic pocket pocket factor Hepatitis B virus / VIRUS LIKE PARTICLE | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmicrotubule-dependent intracellular transport of viral material towards nucleus / T=4 icosahedral viral capsid / viral penetration into host nucleus / host cell / host cell cytoplasm / symbiont entry into host cell / structural molecule activity / DNA binding / RNA binding Similarity search - Function | ||||||||||||||||||
| Biological species |  Hepatitis B virus ayw/France/Tiollais/1979 / Hepatitis B virus ayw/France/Tiollais/1979 /   Hepatitis B virus Hepatitis B virus | ||||||||||||||||||
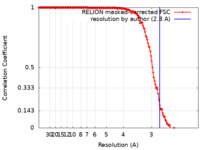
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | ||||||||||||||||||
 Authors Authors | Makbul C / Khayenko V / Maric MH / Bottcher B | ||||||||||||||||||
| Funding support |  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2025 Journal: Elife / Year: 2025Title: Induction of hepatitis B core protein aggregation targeting an unconventional binding site Authors: Khayenko V / Makbul C / Schulte C / Hemmelmann N / Kachler S / Bottcher B / Maric HM / Comas-Garcia M / Dotsch V | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17996.map.gz emd_17996.map.gz | 301 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17996-v30.xml emd-17996-v30.xml emd-17996.xml emd-17996.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17996_fsc.xml emd_17996_fsc.xml | 15.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_17996.png emd_17996.png | 259.6 KB | ||
| Masks |  emd_17996_msk_1.map emd_17996_msk_1.map | 325 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17996.cif.gz emd-17996.cif.gz | 6.4 KB | ||
| Others |  emd_17996_half_map_1.map.gz emd_17996_half_map_1.map.gz emd_17996_half_map_2.map.gz emd_17996_half_map_2.map.gz | 259.2 MB 259.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17996 http://ftp.pdbj.org/pub/emdb/structures/EMD-17996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17996 | HTTPS FTP |
-Related structure data
| Related structure data |  8pwoMC  8px3C  8px6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17996.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17996.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0635 Å | ||||||||||||||||||||||||||||||||||||
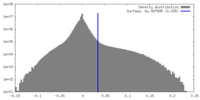
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17996_msk_1.map emd_17996_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
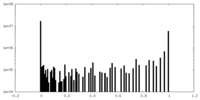
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17996_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
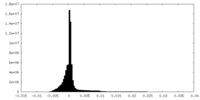
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17996_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
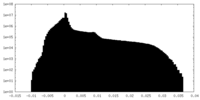
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Hepatitis B virus
| Entire | Name:   Hepatitis B virus Hepatitis B virus |
|---|---|
| Components |
|
-Supramolecule #1: Hepatitis B virus
| Supramolecule | Name: Hepatitis B virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 10407 / Sci species name: Hepatitis B virus / Sci species strain: ayw/France/Tiollais/1979 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 5 MDa |
| Virus shell | Shell ID: 1 / Diameter: 360.0 Å / T number (triangulation number): 4 |
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Details: capsid has bound geraniol / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Hepatitis B virus ayw/France/Tiollais/1979 Hepatitis B virus ayw/France/Tiollais/1979 |
| Molecular weight | Theoretical: 21.146217 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDIDPYKEFG ATVELLSFLP SDFFPSVRDL LDTASALYRE ALESPEHCSP HHTALRQAIL CWGELMTLAT WVGVNLEDPA SRDLVVSYV NTNMGLKFRQ LLWFHISCLT FGRETVIEYL VSFGVWIRTP PAYRPPNAPI LSTLPETTVV RRRGRSPRRR T PSPRRRRS QSPRRRRSQS RESQC UniProtKB: Capsid protein |
-Macromolecule #2: Geraniol
| Macromolecule | Name: Geraniol / type: ligand / ID: 2 / Number of copies: 4 / Formula: 64Z |
|---|---|
| Molecular weight | Theoretical: 154.249 Da |
| Chemical component information |  ChemComp-64Z: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 120 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.029 kPa Details: plasma cleaner (model PDC-002. Harrick Plasma, Ithaca, NY, USA); at medium power of the instrument |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 4875 / Average exposure time: 3.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8pwo: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)