+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of Giardia intestinalis deoxyadenosine kinase | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | kinase / TRANSFERASE | |||||||||||||||
| Biological species |  Giardia intestinalis (eukaryote) Giardia intestinalis (eukaryote) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.304 Å | |||||||||||||||
 Authors Authors | Ranjbarian F / Rafie K / Shankar K / Krakovka S / Svaerd S / Carlson L-A / Hofer A | |||||||||||||||
| Funding support |  Sweden, 4 items Sweden, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2024 Journal: Nucleic Acids Res / Year: 2024Title: Tetramerization of deoxyadenosine kinase meets the demands of a DNA replication substrate challenge in Giardia intestinalis. Authors: Farahnaz Ranjbarian / Karim Rafie / Kasturika Shankar / Sascha Krakovka / Staffan G Svärd / Lars-Anders Carlson / Anders Hofer /   Abstract: The protozoan parasite Giardia intestinalis is one of only a few organisms lacking de novo synthesis of DNA building blocks (deoxyribonucleotides). Instead, the parasite relies exclusively on ...The protozoan parasite Giardia intestinalis is one of only a few organisms lacking de novo synthesis of DNA building blocks (deoxyribonucleotides). Instead, the parasite relies exclusively on salvaging deoxyadenosine and other deoxyribonucleosides from its host environment. Here, we report that G. intestinalis has a deoxyribonucleoside kinase with a 1000-fold higher catalytic efficiency (kcat/KM) for deoxyadenosine than the corresponding mammalian kinases and can thereby provide sufficient deoxyadenosine triphosphate levels for DNA synthesis despite the lack of de novo synthesis. Several deoxyadenosine analogs were also potent substrates and showed comparable EC50 values on cultured G. intestinalis cells as metronidazole, the current first-line treatment, with the additional advantage of being effective against metronidazole-resistant parasites. Structural analysis using cryo-EM and X-ray crystallography showed that the enzyme is unique within its family of deoxyribonucleoside kinases by forming a tetramer stabilized by extended N- and C-termini in a novel dimer-dimer interaction. Removal of the two termini resulted in lost ability to form tetramers and a markedly reduced affinity for the deoxyribonucleoside substrate. The development of highly efficient deoxyribonucleoside kinases via oligomerization may represent a critical evolutionary adaptation in organisms that rely solely on deoxyribonucleoside salvage. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17948.map.gz emd_17948.map.gz | 45.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17948-v30.xml emd-17948-v30.xml emd-17948.xml emd-17948.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17948.png emd_17948.png | 58.9 KB | ||
| Filedesc metadata |  emd-17948.cif.gz emd-17948.cif.gz | 4.8 KB | ||
| Others |  emd_17948_half_map_1.map.gz emd_17948_half_map_1.map.gz emd_17948_half_map_2.map.gz emd_17948_half_map_2.map.gz | 46.2 MB 46.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17948 http://ftp.pdbj.org/pub/emdb/structures/EMD-17948 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17948 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17948 | HTTPS FTP |
-Validation report
| Summary document |  emd_17948_validation.pdf.gz emd_17948_validation.pdf.gz | 165.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17948_full_validation.pdf.gz emd_17948_full_validation.pdf.gz | 165 KB | Display | |
| Data in XML |  emd_17948_validation.xml.gz emd_17948_validation.xml.gz | 573 B | Display | |
| Data in CIF |  emd_17948_validation.cif.gz emd_17948_validation.cif.gz | 486 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17948 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17948 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17948 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17948 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17948.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17948.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.58 Å | ||||||||||||||||||||||||||||||||||||
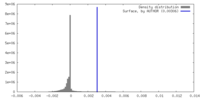
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_17948_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
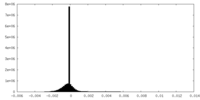
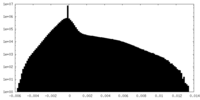
| Density Histograms |
-Half map: #2
| File | emd_17948_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
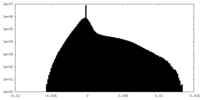
| Density Histograms |
- Sample components
Sample components
-Entire : deoxyadenosine kinase
| Entire | Name: deoxyadenosine kinase |
|---|---|
| Components |
|
-Supramolecule #1: deoxyadenosine kinase
| Supramolecule | Name: deoxyadenosine kinase / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Giardia intestinalis (eukaryote) Giardia intestinalis (eukaryote) |
| Molecular weight | Theoretical: 117 kDa/nm |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4000 pixel / Digitization - Dimensions - Height: 4000 pixel / Number real images: 3420 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 215000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)