+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
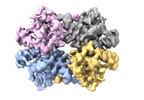
| Title | Cryo-EM structure of Enterobacter sp. 638 BcsD | |||||||||
 Map data Map data | Sharpened cryo-EM map of Enterobacter sp. 638 BcsD | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cellulose secretion / bacterial biofilms / cytoskeleton / CYTOSOLIC PROTEIN | |||||||||
| Function / homology | Cellulose synthase operon protein D, bacterial / Cellulose synthase subunit D superfamily / Cellulose synthase subunit D / cellulose biosynthetic process / Cellulose synthase Function and homology information Function and homology information | |||||||||
| Biological species |  Enterobacter sp. 638 (bacteria) Enterobacter sp. 638 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.15 Å | |||||||||
 Authors Authors | Notopoulou A / Krasteva PV | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Curr Biol / Year: 2024 Journal: Curr Biol / Year: 2024Title: Structures and roles of BcsD and partner scaffold proteins in proteobacterial cellulose secretion. Authors: Thibault G Sana / Areti Notopoulou / Lucie Puygrenier / Marion Decossas / Sandra Moreau / Aurélien Carlier / Petya V Krasteva /   Abstract: Cellulose is the world's most abundant biopolymer, and similar to its role as a cell wall component in plants, it is a prevalent constituent of the extracellular matrix in bacterial biofilms. ...Cellulose is the world's most abundant biopolymer, and similar to its role as a cell wall component in plants, it is a prevalent constituent of the extracellular matrix in bacterial biofilms. Although bacterial cellulose (BC) was first described in the 19 century, it was only recently revealed that it is produced by several distinct types of Bcs secretion systems that feature multiple accessory subunits in addition to a catalytic BcsAB synthase tandem. We recently showed that crystalline cellulose secretion in the Gluconacetobacter genus (α-Proteobacteria) is driven by a supramolecular BcsH-BcsD scaffold-the "cortical belt"-which stabilizes the synthase nanoarrays through an unexpected inside-out mechanism for secretion system assembly. Interestingly, while bcsH is specific for Gluconacetobacter, bcsD homologs are widespread in Proteobacteria. Here, we examine BcsD homologs and their gene neighborhoods from several plant-colonizing β- and γ-Proteobacteria proposed to secrete a variety of non-crystalline and/or chemically modified cellulosic polymers. We provide structural and mechanistic evidence that through different quaternary structure assemblies BcsD acts with proline-rich BcsH, BcsP, or BcsO partners across the proteobacterial clade to form synthase-interacting intracellular scaffolds that, in turn, determine the biofilm strength and architecture in species with strikingly different physiology and secreted biopolymers. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17791.map.gz emd_17791.map.gz | 78.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17791-v30.xml emd-17791-v30.xml emd-17791.xml emd-17791.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17791.png emd_17791.png | 102.6 KB | ||
| Masks |  emd_17791_msk_1.map emd_17791_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17791.cif.gz emd-17791.cif.gz | 6 KB | ||
| Others |  emd_17791_additional_1.map.gz emd_17791_additional_1.map.gz emd_17791_additional_2.map.gz emd_17791_additional_2.map.gz emd_17791_half_map_1.map.gz emd_17791_half_map_1.map.gz emd_17791_half_map_2.map.gz emd_17791_half_map_2.map.gz | 74.1 MB 40.5 MB 77.4 MB 77.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17791 http://ftp.pdbj.org/pub/emdb/structures/EMD-17791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17791 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17791 | HTTPS FTP |
-Related structure data
| Related structure data |  8pogMC  8pkdC  8pocC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17791.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17791.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened cryo-EM map of Enterobacter sp. 638 BcsD | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
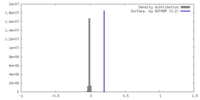
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17791_msk_1.map emd_17791_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
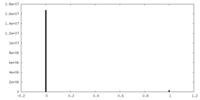
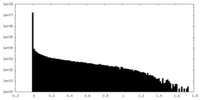
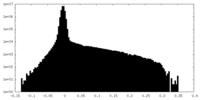
| Density Histograms |
-Additional map: Deep EMhancer sharpened cryo-EM map of Enterobacter sp. 638 BcsD
| File | emd_17791_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Deep EMhancer sharpened cryo-EM map of Enterobacter sp. 638 BcsD | ||||||||||||
| Projections & Slices |
| ||||||||||||
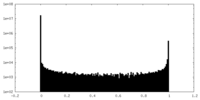
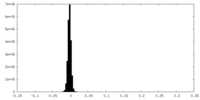
| Density Histograms |
-Additional map: Unsharpened cryo-EM map of Enterobacter sp. 638 BcsD
| File | emd_17791_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened cryo-EM map of Enterobacter sp. 638 BcsD | ||||||||||||
| Projections & Slices |
| ||||||||||||
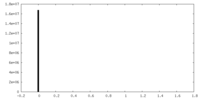
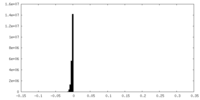
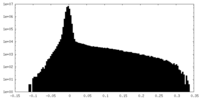
| Density Histograms |
-Half map: Cryo-EM half map for Enterobacter sp. 638 BcsD
| File | emd_17791_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM half map for Enterobacter sp. 638 BcsD | ||||||||||||
| Projections & Slices |
| ||||||||||||
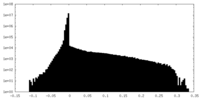
| Density Histograms |
-Half map: Cryo-EM half map for Enterobacter sp. 638 BcsD
| File | emd_17791_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM half map for Enterobacter sp. 638 BcsD | ||||||||||||
| Projections & Slices |
| ||||||||||||
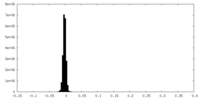
| Density Histograms |
- Sample components
Sample components
-Entire : Tetrameric BcsD of Enterobacter sp. 638
| Entire | Name: Tetrameric BcsD of Enterobacter sp. 638 |
|---|---|
| Components |
|
-Supramolecule #1: Tetrameric BcsD of Enterobacter sp. 638
| Supramolecule | Name: Tetrameric BcsD of Enterobacter sp. 638 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Enterobacter sp. 638 (bacteria) Enterobacter sp. 638 (bacteria) |
| Molecular weight | Theoretical: 70 KDa |
-Macromolecule #1: BcsD of Enterobacter sp. 638
| Macromolecule | Name: BcsD of Enterobacter sp. 638 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Enterobacter sp. 638 (bacteria) Enterobacter sp. 638 (bacteria) |
| Molecular weight | Theoretical: 18.138383 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSTTTLENP ALLGWFRGQQ TPEGWFDLLA LIVDGMVRNV GELESQPFLR QMGIALADQY PLPASETVGE LEANINAQLA RFGWGCVEI DTSDTDLTLR HQALPVSRVE DQQSRWCHAF CAILEGLYVR WMQGQGGKSH VTVSRERLFS LSDVQFRYHN P Q UniProtKB: Cellulose synthase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20 mM HEPES pH 8.0, 120 mM NaCl |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 0-50 / Average electron dose: 49.2 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.4 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Details | Reiterative refinement in Phenix and Coot |
| Refinement | Space: REAL / Protocol: OTHER |
| Output model |  PDB-8pog: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)