[English] 日本語
 Yorodumi
Yorodumi- EMDB-17785: Cryo-EM structure of styrene oxide isomerase bound to benzylamine... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of styrene oxide isomerase bound to benzylamine inhibitor | |||||||||
 Map data Map data | full map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Heme binding protein / Enzyme / Isomerase / MEMBRANE PROTEIN | |||||||||
| Function / homology | : / isomerase activity / membrane / Styrene oxide isomerase Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas sp. VLB120 (bacteria) / Pseudomonas sp. VLB120 (bacteria) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.12 Å | |||||||||
 Authors Authors | Khanppnavar B / Korkhov V / Li X | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem / Year: 2024 Journal: Nat Chem / Year: 2024Title: Structural basis of the Meinwald rearrangement catalysed by styrene oxide isomerase. Authors: Basavraj Khanppnavar / Joel P S Choo / Peter-Leon Hagedoorn / Grigory Smolentsev / Saša Štefanić / Selvapravin Kumaran / Dirk Tischler / Fritz K Winkler / Volodymyr M Korkhov / Zhi Li / ...Authors: Basavraj Khanppnavar / Joel P S Choo / Peter-Leon Hagedoorn / Grigory Smolentsev / Saša Štefanić / Selvapravin Kumaran / Dirk Tischler / Fritz K Winkler / Volodymyr M Korkhov / Zhi Li / Richard A Kammerer / Xiaodan Li /     Abstract: Membrane-bound styrene oxide isomerase (SOI) catalyses the Meinwald rearrangement-a Lewis-acid-catalysed isomerization of an epoxide to a carbonyl compound-and has been used in single and cascade ...Membrane-bound styrene oxide isomerase (SOI) catalyses the Meinwald rearrangement-a Lewis-acid-catalysed isomerization of an epoxide to a carbonyl compound-and has been used in single and cascade reactions. However, the structural information that explains its reaction mechanism has remained elusive. Here we determine cryo-electron microscopy (cryo-EM) structures of SOI bound to a single-domain antibody with and without the competitive inhibitor benzylamine, and elucidate the catalytic mechanism using electron paramagnetic resonance spectroscopy, functional assays, biophysical methods and docking experiments. We find ferric haem b bound at the subunit interface of the trimeric enzyme through H58, where Fe(III) acts as the Lewis acid by binding to the epoxide oxygen. Y103 and N64 and a hydrophobic pocket binding the oxygen of the epoxide and the aryl group, respectively, position substrates in a manner that explains the high regio-selectivity and stereo-specificity of SOI. Our findings can support extending the range of epoxide substrates and be used to potentially repurpose SOI for the catalysis of new-to-nature Fe-based chemical reactions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17785.map.gz emd_17785.map.gz | 21.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17785-v30.xml emd-17785-v30.xml emd-17785.xml emd-17785.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17785_fsc.xml emd_17785_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_17785.png emd_17785.png | 38.3 KB | ||
| Masks |  emd_17785_msk_1.map emd_17785_msk_1.map | 347.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17785.cif.gz emd-17785.cif.gz | 6.5 KB | ||
| Others |  emd_17785_half_map_1.map.gz emd_17785_half_map_1.map.gz emd_17785_half_map_2.map.gz emd_17785_half_map_2.map.gz | 277.8 MB 277.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17785 http://ftp.pdbj.org/pub/emdb/structures/EMD-17785 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17785 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17785 | HTTPS FTP |
-Validation report
| Summary document |  emd_17785_validation.pdf.gz emd_17785_validation.pdf.gz | 767 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17785_full_validation.pdf.gz emd_17785_full_validation.pdf.gz | 766.5 KB | Display | |
| Data in XML |  emd_17785_validation.xml.gz emd_17785_validation.xml.gz | 23.4 KB | Display | |
| Data in CIF |  emd_17785_validation.cif.gz emd_17785_validation.cif.gz | 31.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17785 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17785 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17785 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17785 | HTTPS FTP |
-Related structure data
| Related structure data |  8pnuMC  8pnvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17785.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17785.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.66 Å | ||||||||||||||||||||||||||||||||||||
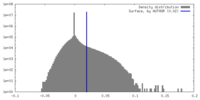
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17785_msk_1.map emd_17785_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half1
| File | emd_17785_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half2
| File | emd_17785_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Styrene oxide isomerase-Nanobody-Benzylamine complex
| Entire | Name: Styrene oxide isomerase-Nanobody-Benzylamine complex |
|---|---|
| Components |
|
-Supramolecule #1: Styrene oxide isomerase-Nanobody-Benzylamine complex
| Supramolecule | Name: Styrene oxide isomerase-Nanobody-Benzylamine complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Styrene oxide isomerase bound to nanobody complex and benzylamine inhibitor |
|---|---|
| Source (natural) | Organism:  Pseudomonas sp. VLB120 (bacteria) Pseudomonas sp. VLB120 (bacteria) |
| Molecular weight | Theoretical: 201.7 kDa/nm |
-Macromolecule #1: Styrene oxide isomerase
| Macromolecule | Name: Styrene oxide isomerase / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas sp. VLB120 (bacteria) / Strain: Pseudomonas sp. VLB120 / Cell: bacteria Pseudomonas sp. VLB120 (bacteria) / Strain: Pseudomonas sp. VLB120 / Cell: bacteria |
| Molecular weight | Theoretical: 19.680867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SQDPMLHAFE RKMAGHGILM IFCTLLFGVG LWMNLVGGFE IIPGYIIEFH VPGSPEGWAR AHSGPALNGM MVIAVAFVL PSLGFADKTA RLLGSIIVLD GWSNVGFYLF SNFSPNRGLT FGPNQFGPGD IFSFLALAPA YLFGVLAMGA L AVIGYQAL KSTRSRKAVP HAAAE UniProtKB: Styrene oxide isomerase |
-Macromolecule #2: Nanobody
| Macromolecule | Name: Nanobody / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.055473 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AQGQLVESGG GLVQAGGSLR LSCTGSGRAF VTPAVGWFRQ APGKEREFVG TINWSGSHTS YADPVKGRFT ISRDNAKETV YLQMNNLKP EDADVYYCAS RGVSGRYEYW GKGTPVTVSS HHHHHHEPEA |
-Macromolecule #3: BENZYLAMINE
| Macromolecule | Name: BENZYLAMINE / type: ligand / ID: 3 / Number of copies: 6 / Formula: ABN |
|---|---|
| Molecular weight | Theoretical: 107.153 Da |
| Chemical component information |  ChemComp-ABN: |
-Macromolecule #4: PROTOPORPHYRIN IX CONTAINING FE
| Macromolecule | Name: PROTOPORPHYRIN IX CONTAINING FE / type: ligand / ID: 4 / Number of copies: 6 / Formula: HEM |
|---|---|
| Molecular weight | Theoretical: 616.487 Da |
| Chemical component information |  ChemComp-HEM: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 264 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)