[English] 日本語
 Yorodumi
Yorodumi- EMDB-17779: Structure of human oligosaccharyltransferase OST-A complex bound ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
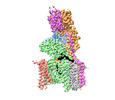
| Title | Structure of human oligosaccharyltransferase OST-A complex bound to NGI-1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | N-glycosylation / OST-A complex / NGI-1 inhibitor / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationoligosaccharyltransferase complex binding / oligosaccharyltransferase complex A / oligosaccharyltransferase complex B / Asparagine N-linked glycosylation / dolichyl-diphosphooligosaccharide-protein glycotransferase / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / oligosaccharyltransferase complex / protein N-linked glycosylation via asparagine / : / protein N-linked glycosylation ...oligosaccharyltransferase complex binding / oligosaccharyltransferase complex A / oligosaccharyltransferase complex B / Asparagine N-linked glycosylation / dolichyl-diphosphooligosaccharide-protein glycotransferase / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / oligosaccharyltransferase complex / protein N-linked glycosylation via asparagine / : / protein N-linked glycosylation / epithelial cell apoptotic process / azurophil granule membrane / : / Advanced glycosylation endproduct receptor signaling / blastocyst development / SRP-dependent cotranslational protein targeting to membrane / rough endoplasmic reticulum / response to cytokine / post-translational protein modification / response to endoplasmic reticulum stress / T cell activation / enzyme activator activity / regulation of protein stability / protein modification process / melanosome / protein-macromolecule adaptor activity / Maturation of spike protein / nuclear body / inflammatory response / intracellular membrane-bounded organelle / apoptotic process / Neutrophil degranulation / endoplasmic reticulum membrane / negative regulation of apoptotic process / endoplasmic reticulum / RNA binding / metal ion binding / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.61 Å | |||||||||
 Authors Authors | Ramirez AS / Kowal J / Locher KP | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: Positive selection CRISPR screens reveal a druggable pocket in an oligosaccharyltransferase required for inflammatory signaling to NF-κB. Authors: Benjamin L Lampson / Ana S Ramίrez / Marta Baro / Lixia He / Mudra Hegde / Vidyasagar Koduri / Jamie L Pfaff / Ruth E Hanna / Julia Kowal / Nitin H Shirole / Yanfeng He / John G Doench / ...Authors: Benjamin L Lampson / Ana S Ramίrez / Marta Baro / Lixia He / Mudra Hegde / Vidyasagar Koduri / Jamie L Pfaff / Ruth E Hanna / Julia Kowal / Nitin H Shirole / Yanfeng He / John G Doench / Joseph N Contessa / Kaspar P Locher / William G Kaelin /   Abstract: Nuclear factor κB (NF-κB) plays roles in various diseases. Many inflammatory signals, such as circulating lipopolysaccharides (LPSs), activate NF-κB via specific receptors. Using whole-genome ...Nuclear factor κB (NF-κB) plays roles in various diseases. Many inflammatory signals, such as circulating lipopolysaccharides (LPSs), activate NF-κB via specific receptors. Using whole-genome CRISPR-Cas9 screens of LPS-treated cells that express an NF-κB-driven suicide gene, we discovered that the LPS receptor Toll-like receptor 4 (TLR4) is specifically dependent on the oligosaccharyltransferase complex OST-A for N-glycosylation and cell-surface localization. The tool compound NGI-1 inhibits OST complexes in vivo, but the underlying molecular mechanism remained unknown. We did a CRISPR base-editor screen for NGI-1-resistant variants of STT3A, the catalytic subunit of OST-A. These variants, in conjunction with cryoelectron microscopy studies, revealed that NGI-1 binds the catalytic site of STT3A, where it traps a molecule of the donor substrate dolichyl-PP-GlcNAc-Man-Glc, suggesting an uncompetitive inhibition mechanism. Our results provide a rationale for and an initial step toward the development of STT3A-specific inhibitors and illustrate the power of contemporaneous base-editor and structural studies to define drug mechanism of action. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17779.map.gz emd_17779.map.gz | 483.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17779-v30.xml emd-17779-v30.xml emd-17779.xml emd-17779.xml | 25.8 KB 25.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17779.png emd_17779.png | 86.8 KB | ||
| Filedesc metadata |  emd-17779.cif.gz emd-17779.cif.gz | 8.6 KB | ||
| Others |  emd_17779_half_map_1.map.gz emd_17779_half_map_1.map.gz emd_17779_half_map_2.map.gz emd_17779_half_map_2.map.gz | 475.7 MB 475.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17779 http://ftp.pdbj.org/pub/emdb/structures/EMD-17779 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17779 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17779 | HTTPS FTP |
-Validation report
| Summary document |  emd_17779_validation.pdf.gz emd_17779_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17779_full_validation.pdf.gz emd_17779_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_17779_validation.xml.gz emd_17779_validation.xml.gz | 18.5 KB | Display | |
| Data in CIF |  emd_17779_validation.cif.gz emd_17779_validation.cif.gz | 21.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17779 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17779 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17779 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17779 | HTTPS FTP |
-Related structure data
| Related structure data |  8pn9MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17779.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17779.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.648 Å | ||||||||||||||||||||||||||||||||||||
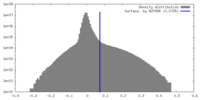
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_17779_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17779_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : OLIGOSACCHARYLTRANSFERASE COMPLEX OST-A WITH BOUND NGI-1 INHIBITOR
+Supramolecule #1: OLIGOSACCHARYLTRANSFERASE COMPLEX OST-A WITH BOUND NGI-1 INHIBITOR
+Macromolecule #1: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #2: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #3: Transmembrane protein 258
+Macromolecule #4: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #5: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #6: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #7: Oligosaccharyltransferase complex subunit OSTC
+Macromolecule #8: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48...
+Macromolecule #13: (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-[(1~{S},2~{R}...
+Macromolecule #14: (4R,7R)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(undecanoyloxy)met...
+Macromolecule #15: MANGANESE (II) ION
+Macromolecule #16: (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRI...
+Macromolecule #17: 5-(dimethylsulfamoyl)-~{N}-(5-methyl-1,3-thiazol-2-yl)-2-pyrrolid...
+Macromolecule #18: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 54.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.61 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 101699 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)