+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of NR5A2-nucleosome complex SHL+5.5 | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | Nucleosome / nuclear receptor / NR5A2 / DNA BINDING PROTEIN | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報Deposition of new CENPA-containing nucleosomes at the centromere / Inhibition of DNA recombination at telomere / SUMOylation of chromatin organization proteins / E3 ubiquitin ligases ubiquitinate target proteins / positive regulation of glucocorticoid biosynthetic process / zygotic genome activation / positive regulation of tendon cell differentiation / DNA Damage/Telomere Stress Induced Senescence / morula formation / Regulation of gene expression in early pancreatic precursor cells ...Deposition of new CENPA-containing nucleosomes at the centromere / Inhibition of DNA recombination at telomere / SUMOylation of chromatin organization proteins / E3 ubiquitin ligases ubiquitinate target proteins / positive regulation of glucocorticoid biosynthetic process / zygotic genome activation / positive regulation of tendon cell differentiation / DNA Damage/Telomere Stress Induced Senescence / morula formation / Regulation of gene expression in early pancreatic precursor cells / G2/M DNA damage checkpoint / primary ovarian follicle growth / HDMs demethylate histones / Regulation of endogenous retroelements by KRAB-ZFP proteins / Condensation of Prophase Chromosomes / Nonhomologous End-Joining (NHEJ) / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine / HDACs deacetylate histones / pancreas morphogenesis / PRC2 methylates histones and DNA / Processing of DNA double-strand break ends / HATs acetylate histones / MLL4 and MLL3 complexes regulate expression of PPARG target genes in adipogenesis and hepatic steatosis / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / PKMTs methylate histone lysines / inner cell mass cell differentiation / tissue development / RUNX1 regulates genes involved in megakaryocyte differentiation and platelet function / acinar cell differentiation / RMTs methylate histone arginines / negative regulation of chromosome condensation / Sertoli cell development / positive regulation of stem cell differentiation / Barr body / : / Factors involved in megakaryocyte development and platelet production / positive regulation of T cell anergy / Estrogen-dependent gene expression / embryonic cleavage / pericentric heterochromatin formation / inner kinetochore / bile acid metabolic process / embryo development ending in birth or egg hatching / muscle cell differentiation / exocrine pancreas development / cartilage development / negative regulation of chondrocyte differentiation / oocyte maturation / nucleosomal DNA binding / oogenesis / homeostatic process / nucleus organization / calcineurin-mediated signaling / chromosome, centromeric region / somatic stem cell population maintenance / spermatid development / single fertilization / subtelomeric heterochromatin formation / positive regulation of viral genome replication / RNA polymerase II core promoter sequence-specific DNA binding / protein localization to CENP-A containing chromatin / CENP-A containing nucleosome / hormone-mediated signaling pathway / neurogenesis / positive regulation of T cell proliferation / embryo implantation / cholesterol homeostasis / cellular response to leukemia inhibitory factor / transcription coregulator binding / SUMOylation of intracellular receptors / phospholipid binding / positive regulation of T cell activation / Nuclear Receptor transcription pathway / negative regulation of inflammatory response / RNA polymerase II transcription regulator complex / multicellular organism growth / male gonad development / nuclear receptor activity / sequence-specific double-stranded DNA binding / osteoblast differentiation / structural constituent of chromatin / nucleosome / nucleosome assembly / chromosome / chromatin organization / positive regulation of cell growth / DNA-binding transcription activator activity, RNA polymerase II-specific / spermatogenesis / Estrogen-dependent gene expression / sequence-specific DNA binding / DNA-binding transcription factor activity, RNA polymerase II-specific / chromosome, telomeric region / cell population proliferation / transcription cis-regulatory region binding / RNA polymerase II cis-regulatory region sequence-specific DNA binding / chromatin remodeling / DNA-binding transcription factor activity / protein heterodimerization activity / chromatin binding 類似検索 - 分子機能 | |||||||||
| 生物種 |   Homo sapiens (ヒト) / synthetic construct (人工物) Homo sapiens (ヒト) / synthetic construct (人工物) | |||||||||
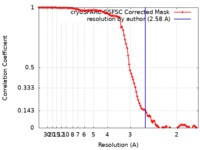
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.58 Å | |||||||||
 データ登録者 データ登録者 | Kobayashi W / Sappler A / Bollschweiler D / Kummecke M / Basquin J / Arslantas E / Ruangroengkulrith S / Hornberger R / Duderstadt K / Tachibana K | |||||||||
| 資金援助 |  ドイツ, European Union, 2件 ドイツ, European Union, 2件
| |||||||||
 引用 引用 |  ジャーナル: Nat Struct Mol Biol / 年: 2024 ジャーナル: Nat Struct Mol Biol / 年: 2024タイトル: Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition. 著者: Wataru Kobayashi / Anna H Sappler / Daniel Bollschweiler / Maximilian Kümmecke / Jérôme Basquin / Eda Nur Arslantas / Siwat Ruangroengkulrith / Renate Hornberger / Karl Duderstadt / Kikuë Tachibana /  要旨: Gene expression during natural and induced reprogramming is controlled by pioneer transcription factors that initiate transcription from closed chromatin. Nr5a2 is a key pioneer factor that regulates ...Gene expression during natural and induced reprogramming is controlled by pioneer transcription factors that initiate transcription from closed chromatin. Nr5a2 is a key pioneer factor that regulates zygotic genome activation in totipotent embryos, pluripotency in embryonic stem cells and metabolism in adult tissues, but the mechanism of its pioneer activity remains poorly understood. Here, we present a cryo-electron microscopy structure of human NR5A2 bound to a nucleosome. The structure shows that the conserved carboxy-terminal extension (CTE) loop of the NR5A2 DNA-binding domain competes with a DNA minor groove anchor of the nucleosome and releases entry-exit site DNA. Mutational analysis showed that NR5A2 D159 of the CTE is dispensable for DNA binding but required for stable nucleosome association and persistent DNA 'unwrapping'. These findings suggest that NR5A2 belongs to an emerging class of pioneer factors that can use DNA minor groove anchor competition to destabilize nucleosomes and facilitate gene expression during reprogramming. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_17740.map.gz emd_17740.map.gz | 63 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-17740-v30.xml emd-17740-v30.xml emd-17740.xml emd-17740.xml | 21.2 KB 21.2 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_17740_fsc.xml emd_17740_fsc.xml | 10.5 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_17740.png emd_17740.png | 70.2 KB | ||
| Filedesc metadata |  emd-17740.cif.gz emd-17740.cif.gz | 6.5 KB | ||
| その他 |  emd_17740_half_map_1.map.gz emd_17740_half_map_1.map.gz emd_17740_half_map_2.map.gz emd_17740_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17740 http://ftp.pdbj.org/pub/emdb/structures/EMD-17740 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17740 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17740 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_17740_validation.pdf.gz emd_17740_validation.pdf.gz | 1 MB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_17740_full_validation.pdf.gz emd_17740_full_validation.pdf.gz | 1 MB | 表示 | |
| XML形式データ |  emd_17740_validation.xml.gz emd_17740_validation.xml.gz | 19.1 KB | 表示 | |
| CIF形式データ |  emd_17740_validation.cif.gz emd_17740_validation.cif.gz | 24.5 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17740 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17740 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17740 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17740 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8pkiMC  8pkjC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_17740.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_17740.map.gz / 形式: CCP4 / 大きさ: 125 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.8512 Å | ||||||||||||||||||||||||||||||||||||
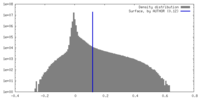
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: #2
| ファイル | emd_17740_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
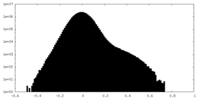
| 密度ヒストグラム |
-ハーフマップ: #1
| ファイル | emd_17740_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
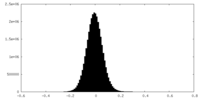
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Cryo-EM structure of the nucleosome containing NR5A2 motif at SHL+5.5
| 全体 | 名称: Cryo-EM structure of the nucleosome containing NR5A2 motif at SHL+5.5 |
|---|---|
| 要素 |
|
-超分子 #1: Cryo-EM structure of the nucleosome containing NR5A2 motif at SHL+5.5
| 超分子 | 名称: Cryo-EM structure of the nucleosome containing NR5A2 motif at SHL+5.5 タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#7 |
|---|---|
| 由来(天然) | 生物種:  |
-分子 #1: Histone H3.3
| 分子 | 名称: Histone H3.3 / タイプ: protein_or_peptide / ID: 1 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 15.360983 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MARTKQTARK STGGKAPRKQ LATKAARKSA PSTGGVKKPH RYRPGTVALR EIRRYQKSTE LLIRKLPFQR LVREIAQDFK TDLRFQSAA IGALQEASEA YLVGLFEDTN LCAIHAKRVT IMPKDIQLAR RIRGERA UniProtKB: Histone H3.3 |
-分子 #2: Histone H4
| 分子 | 名称: Histone H4 / タイプ: protein_or_peptide / ID: 2 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 11.394426 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK VFLENVIRDA VTYTEHAKRK TVTAMDVVY ALKRQGRTLY GFGG UniProtKB: Histone H4 |
-分子 #3: Histone H2A
| 分子 | 名称: Histone H2A / タイプ: protein_or_peptide / ID: 3 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 14.165551 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MSGRGKQGGK ARAKAKTRSS RAGLQFPVGR VHRLLRKGNY SERVGAGAPV YLAAVLEYLT AEILELAGNA ARDNKKTRII PRHLQLAIR NDEELNKLLG RVTIAQGGVL PNIQAVLLPK KTESHHKAKG K UniProtKB: Histone H2A |
-分子 #4: Histone H2B type 1-C/E/G
| 分子 | 名称: Histone H2B type 1-C/E/G / タイプ: protein_or_peptide / ID: 4 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 13.937213 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: MPEPAKSAPA PKKGSKKAVT KAQKKDGKKR KRSRKESYSV YVYKVLKQVH PDTGISSKAM GIMNSFVNDI FERIAGEASR LAHYNKRST ITSREIQTAV RLLLPGELAK HAVSEGTKAV TKYTSSK UniProtKB: Histone H2B type 1-C/E/G |
-分子 #7: Nuclear receptor subfamily 5 group A member 2
| 分子 | 名称: Nuclear receptor subfamily 5 group A member 2 / タイプ: protein_or_peptide / ID: 7 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 10.989939 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: LCPVCGDKVS GYHYGLLTCE SCKGFFKRTV QNNKRYTCIE NQNCQIDKTQ RKRCPYCRFQ KCLSVGMKLE AVRADRMRGG RNKFGPMYK RDRAL UniProtKB: Nuclear receptor subfamily 5 group A member 2 |
-分子 #5: DNA
| 分子 | 名称: DNA / タイプ: dna / ID: 5 / コピー数: 1 / 分類: DNA |
|---|---|
| 由来(天然) | 生物種: synthetic construct (人工物) |
| 分子量 | 理論値: 46.968922 KDa |
| 配列 | 文字列: (DA)(DT)(DC)(DC)(DT)(DG)(DG)(DA)(DG)(DA) (DA)(DT)(DC)(DC)(DC)(DG)(DG)(DT)(DG)(DC) (DC)(DG)(DA)(DG)(DG)(DC)(DC)(DG)(DC) (DT)(DC)(DA)(DA)(DT)(DT)(DG)(DG)(DT)(DC) (DG) (DT)(DA)(DG)(DA)(DC) ...文字列: (DA)(DT)(DC)(DC)(DT)(DG)(DG)(DA)(DG)(DA) (DA)(DT)(DC)(DC)(DC)(DG)(DG)(DT)(DG)(DC) (DC)(DG)(DA)(DG)(DG)(DC)(DC)(DG)(DC) (DT)(DC)(DA)(DA)(DT)(DT)(DG)(DG)(DT)(DC) (DG) (DT)(DA)(DG)(DA)(DC)(DA)(DG)(DC) (DT)(DC)(DT)(DA)(DG)(DC)(DA)(DC)(DC)(DG) (DC)(DT) (DT)(DA)(DA)(DA)(DC)(DG)(DC) (DA)(DC)(DG)(DT)(DA)(DC)(DG)(DC)(DG)(DC) (DT)(DG)(DT) (DC)(DC)(DC)(DC)(DC)(DG) (DC)(DG)(DT)(DT)(DT)(DT)(DA)(DA)(DC)(DC) (DG)(DC)(DC)(DA) (DA)(DG)(DG)(DG)(DG) (DA)(DT)(DT)(DA)(DC)(DT)(DC)(DC)(DC)(DT) (DA)(DG)(DT)(DC)(DT) (DC)(DC)(DA)(DG) (DG)(DC)(DA)(DC)(DG)(DT)(DT)(DC)(DA)(DA) (DG)(DG)(DC)(DC)(DA)(DA) (DT)(DA)(DC) (DA)(DT)(DC)(DC)(DT)(DG)(DT)(DG)(DA)(DT) |
-分子 #6: DNA
| 分子 | 名称: DNA / タイプ: dna / ID: 6 / コピー数: 1 / 分類: DNA |
|---|---|
| 由来(天然) | 生物種: synthetic construct (人工物) |
| 分子量 | 理論値: 47.489234 KDa |
| 配列 | 文字列: (DA)(DT)(DC)(DA)(DC)(DA)(DG)(DG)(DA)(DT) (DG)(DT)(DA)(DT)(DT)(DG)(DG)(DC)(DC)(DT) (DT)(DG)(DA)(DA)(DC)(DG)(DT)(DG)(DC) (DC)(DT)(DG)(DG)(DA)(DG)(DA)(DC)(DT)(DA) (DG) (DG)(DG)(DA)(DG)(DT) ...文字列: (DA)(DT)(DC)(DA)(DC)(DA)(DG)(DG)(DA)(DT) (DG)(DT)(DA)(DT)(DT)(DG)(DG)(DC)(DC)(DT) (DT)(DG)(DA)(DA)(DC)(DG)(DT)(DG)(DC) (DC)(DT)(DG)(DG)(DA)(DG)(DA)(DC)(DT)(DA) (DG) (DG)(DG)(DA)(DG)(DT)(DA)(DA)(DT) (DC)(DC)(DC)(DC)(DT)(DT)(DG)(DG)(DC)(DG) (DG)(DT) (DT)(DA)(DA)(DA)(DA)(DC)(DG) (DC)(DG)(DG)(DG)(DG)(DG)(DA)(DC)(DA)(DG) (DC)(DG)(DC) (DG)(DT)(DA)(DC)(DG)(DT) (DG)(DC)(DG)(DT)(DT)(DT)(DA)(DA)(DG)(DC) (DG)(DG)(DT)(DG) (DC)(DT)(DA)(DG)(DA) (DG)(DC)(DT)(DG)(DT)(DC)(DT)(DA)(DC)(DG) (DA)(DC)(DC)(DA)(DA) (DT)(DT)(DG)(DA) (DG)(DC)(DG)(DG)(DC)(DC)(DT)(DC)(DG)(DG) (DC)(DA)(DC)(DC)(DG)(DG) (DG)(DA)(DT) (DT)(DC)(DT)(DC)(DC)(DA)(DG)(DG)(DA)(DT) |
-分子 #8: ZINC ION
| 分子 | 名称: ZINC ION / タイプ: ligand / ID: 8 / コピー数: 1 / 式: ZN |
|---|---|
| 分子量 | 理論値: 65.409 Da |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 平均電子線量: 65.4 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: SPOT SCAN / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.2 µm / 最小 デフォーカス(公称値): 0.6 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)