+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Escherichia coli paused disome complex (queueing 70S non-rotated closed PRE state) | |||||||||
 マップデータ マップデータ | Cryo-EM reconstruction of the E. coli disome complex (local refinement of queueing 70S non-rotated closed PRE state) | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | translation / disome / collision / PURE system / RIBOSOME | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報positive regulation of cytoplasmic translation / negative regulation of cytoplasmic translational initiation / stringent response / ornithine decarboxylase inhibitor activity / transcription antitermination factor activity, RNA binding / misfolded RNA binding / Group I intron splicing / RNA folding / transcriptional attenuation / endoribonuclease inhibitor activity ...positive regulation of cytoplasmic translation / negative regulation of cytoplasmic translational initiation / stringent response / ornithine decarboxylase inhibitor activity / transcription antitermination factor activity, RNA binding / misfolded RNA binding / Group I intron splicing / RNA folding / transcriptional attenuation / endoribonuclease inhibitor activity / positive regulation of ribosome biogenesis / RNA-binding transcription regulator activity / translational termination / negative regulation of cytoplasmic translation / four-way junction DNA binding / DnaA-L2 complex / translation repressor activity / regulation of mRNA stability / negative regulation of translational initiation / negative regulation of DNA-templated DNA replication initiation / mRNA regulatory element binding translation repressor activity / positive regulation of RNA splicing / assembly of large subunit precursor of preribosome / cytosolic ribosome assembly / regulation of DNA-templated transcription elongation / response to reactive oxygen species / ribosome assembly / transcription elongation factor complex / DNA endonuclease activity / transcription antitermination / regulation of cell growth / translational initiation / DNA-templated transcription termination / response to radiation / maintenance of translational fidelity / mRNA 5'-UTR binding / regulation of translation / large ribosomal subunit / ribosome biogenesis / ribosome binding / transferase activity / ribosomal small subunit biogenesis / ribosomal small subunit assembly / 5S rRNA binding / ribosomal large subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / single-stranded RNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / response to antibiotic / negative regulation of DNA-templated transcription / mRNA binding / DNA binding / RNA binding / zinc ion binding / membrane / cytosol / cytoplasm 類似検索 - 分子機能 | |||||||||
| 生物種 |  | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.3 Å | |||||||||
 データ登録者 データ登録者 | Fluegel T / Schacherl M | |||||||||
| 資金援助 |  ドイツ, 1件 ドイツ, 1件
| |||||||||
 引用 引用 |  ジャーナル: Nat Commun / 年: 2024 ジャーナル: Nat Commun / 年: 2024タイトル: Transient disome complex formation in native polysomes during ongoing protein synthesis captured by cryo-EM. 著者: Timo Flügel / Magdalena Schacherl / Anett Unbehaun / Birgit Schroeer / Marylena Dabrowski / Jörg Bürger / Thorsten Mielke / Thiemo Sprink / Christoph A Diebolder / Yollete V Guillén ...著者: Timo Flügel / Magdalena Schacherl / Anett Unbehaun / Birgit Schroeer / Marylena Dabrowski / Jörg Bürger / Thorsten Mielke / Thiemo Sprink / Christoph A Diebolder / Yollete V Guillén Schlippe / Christian M T Spahn /  要旨: Structural studies of translating ribosomes traditionally rely on in vitro assembly and stalling of ribosomes in defined states. To comprehensively visualize bacterial translation, we reactivated ex ...Structural studies of translating ribosomes traditionally rely on in vitro assembly and stalling of ribosomes in defined states. To comprehensively visualize bacterial translation, we reactivated ex vivo-derived E. coli polysomes in the PURE in vitro translation system and analyzed the actively elongating polysomes by cryo-EM. We find that 31% of 70S ribosomes assemble into disome complexes that represent eight distinct functional states including decoding and termination intermediates, and a pre-nucleophilic attack state. The functional diversity of disome complexes together with RNase digest experiments suggests that paused disome complexes transiently form during ongoing elongation. Structural analysis revealed five disome interfaces between leading and queueing ribosomes that undergo rearrangements as the leading ribosome traverses through the elongation cycle. Our findings reveal at the molecular level how bL9's CTD obstructs the factor binding site of queueing ribosomes to thwart harmful collisions and illustrate how translation dynamics reshape inter-ribosomal contacts. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_17631.map.gz emd_17631.map.gz | 1.1 GB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-17631-v30.xml emd-17631-v30.xml emd-17631.xml emd-17631.xml | 84.4 KB 84.4 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_17631_fsc.xml emd_17631_fsc.xml | 23.2 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_17631.png emd_17631.png | 146 KB | ||
| マスクデータ |  emd_17631_msk_1.map emd_17631_msk_1.map | 1.3 GB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-17631.cif.gz emd-17631.cif.gz | 17.3 KB | ||
| その他 |  emd_17631_half_map_1.map.gz emd_17631_half_map_1.map.gz emd_17631_half_map_2.map.gz emd_17631_half_map_2.map.gz | 1.2 GB 1.2 GB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17631 http://ftp.pdbj.org/pub/emdb/structures/EMD-17631 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17631 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17631 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_17631_validation.pdf.gz emd_17631_validation.pdf.gz | 164.8 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_17631_full_validation.pdf.gz emd_17631_full_validation.pdf.gz | 164.4 KB | 表示 | |
| XML形式データ |  emd_17631_validation.xml.gz emd_17631_validation.xml.gz | 571 B | 表示 | |
| CIF形式データ |  emd_17631_validation.cif.gz emd_17631_validation.cif.gz | 482 B | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17631 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17631 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17631 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17631 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8pegMC  8pklC  8r3vC  8rclC  8rcmC  8rcsC  8rctC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_17631.map.gz / 形式: CCP4 / 大きさ: 1.3 GB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_17631.map.gz / 形式: CCP4 / 大きさ: 1.3 GB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Cryo-EM reconstruction of the E. coli disome complex (local refinement of queueing 70S non-rotated closed PRE state) | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-マスク #1
| ファイル |  emd_17631_msk_1.map emd_17631_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
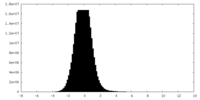
| 密度ヒストグラム |
-ハーフマップ: Cryo-EM reconstruction of the E. coli disome complex...
| ファイル | emd_17631_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Cryo-EM reconstruction of the E. coli disome complex (local refinement of queueing 70S non-rotated closed PRE state. Half map B) | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Cryo-EM reconstruction of the E. coli disome complex...
| ファイル | emd_17631_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Cryo-EM reconstruction of the E. coli disome complex (local refinement of queueing 70S non-rotated closed PRE state. Half map A) | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
+全体 : Escherichia coli paused disome complex
+超分子 #1: Escherichia coli paused disome complex
+分子 #1: 50S ribosomal protein L36
+分子 #2: 50S ribosomal protein L28
+分子 #3: 50S ribosomal protein L29
+分子 #4: 50S ribosomal protein L30
+分子 #5: Large ribosomal subunit protein bL31A
+分子 #6: 50S ribosomal protein L32
+分子 #7: 50S ribosomal protein L35
+分子 #11: Small ribosomal subunit protein uS2
+分子 #12: Small ribosomal subunit protein uS3
+分子 #13: Small ribosomal subunit protein uS4
+分子 #14: Small ribosomal subunit protein uS5
+分子 #15: 30S ribosomal protein S6
+分子 #16: 30S ribosomal protein S7
+分子 #17: Small ribosomal subunit protein uS8
+分子 #18: Small ribosomal subunit protein uS9
+分子 #19: 30S ribosomal protein S10
+分子 #20: Small ribosomal subunit protein uS11
+分子 #21: Small ribosomal subunit protein uS12
+分子 #22: Small ribosomal subunit protein uS13
+分子 #23: Small ribosomal subunit protein uS14
+分子 #24: 30S ribosomal protein S15
+分子 #25: 30S ribosomal protein S16
+分子 #26: Small ribosomal subunit protein uS17
+分子 #27: Small ribosomal subunit protein bS18
+分子 #28: Small ribosomal subunit protein uS19
+分子 #29: Small ribosomal subunit protein bS20
+分子 #30: 30S ribosomal protein S21
+分子 #35: 30S ribosomal protein S1
+分子 #36: Large ribosomal subunit protein uL1
+分子 #37: 50S ribosomal protein L2
+分子 #38: 50S ribosomal protein L3
+分子 #39: 50S ribosomal protein L4
+分子 #40: 50S ribosomal protein L5
+分子 #41: 50S ribosomal protein L6
+分子 #42: 50S ribosomal protein L33
+分子 #43: 50S ribosomal protein L16
+分子 #44: 50S ribosomal protein L9
+分子 #45: Large ribosomal subunit protein uL10
+分子 #46: 50S ribosomal protein L11
+分子 #47: 50S ribosomal protein L34
+分子 #48: 50S ribosomal protein L13
+分子 #49: 50S ribosomal protein L14
+分子 #50: 50S ribosomal protein L15
+分子 #51: Nascent chain
+分子 #52: 50S ribosomal protein L17
+分子 #53: 50S ribosomal protein L18
+分子 #54: 50S ribosomal protein L19
+分子 #55: 50S ribosomal protein L20
+分子 #56: 50S ribosomal protein L21
+分子 #57: 50S ribosomal protein L22
+分子 #58: 50S ribosomal protein L23
+分子 #59: 50S ribosomal protein L24
+分子 #60: 50S ribosomal protein L25
+分子 #61: 50S ribosomal protein L27
+分子 #8: 23S ribosomal RNA
+分子 #9: 5S ribosomal RNA
+分子 #10: 16S ribosomal RNA
+分子 #31: mRNA
+分子 #32: tRNA-Phe (P-site)
+分子 #33: tRNA-Ser (E-site)
+分子 #34: tRNA-Val (A-site)
+分子 #62: ZINC ION
+分子 #63: MAGNESIUM ION
+分子 #64: ADENOSINE-5'-TRIPHOSPHATE
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.6 |
|---|---|
| グリッド | モデル: Quantifoil R2/2 / 材質: COPPER / メッシュ: 300 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: HOLEY / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 20 sec. / 前処理 - 雰囲気: AIR / 前処理 - 気圧: 0.037 kPa 詳細: Operated at 15 mA, easiGlow Discharge Cleaning system (PELCO) |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 75 % / チャンバー内温度: 277 K / 装置: FEI VITROBOT MARK IV 詳細: Withdrawn samples were spotted directly onto freshly glow-discharged holey carbon grids, blotted for 1-2 s, and flash frozen in liquid ethane using a Vitrobot Mark IV plunger (ThermoFisher ...詳細: Withdrawn samples were spotted directly onto freshly glow-discharged holey carbon grids, blotted for 1-2 s, and flash frozen in liquid ethane using a Vitrobot Mark IV plunger (ThermoFisher Scientific) after a wait time of 40 s at 4 degrees Celcius.. |
| 詳細 | In vitro translation reactions were performed in the PURE translation system using the PURExpress delta ribosome kit (NEB, #E3313S). Translation reactions were supplemented with 0.8 U/uL RNAsin Plus RNase Inhibitor (Promega, N261B). SolA, factor mix, and RNAsin Plus were combined on ice, followed by a preincubation at 37 degrees Celcius for 2 min, and added directly to polysomes (700 nM final concentration) that had been preincubated at 37 degrees Celsius for 2 min. After 1 min reaction time, 4 uL of the reaction mixture were withdrawn for plunge freezing. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 温度 | 最低: 80.0 K / 最高: 82.0 K |
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 撮影したグリッド数: 1 / 実像数: 8982 / 平均露光時間: 1.13 sec. / 平均電子線量: 45.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 50.0 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 2.0 µm / 最小 デフォーカス(公称値): 0.5 µm / 倍率(公称値): 81000 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 初期モデル | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| 詳細 | The atomic model of the E. coli 70S ribosome (PDB ID: 7N1P) was initially docked into the postprocessed density maps with UCSF Chimera and manually adjusted in Coot. All models were refined over multiple rounds using the module phenix.real_space_refine in PHENIX and interactive model building and refinement in Coot, using libG restraints for the RNAs. |
| 精密化 | 空間: REAL / プロトコル: RIGID BODY FIT / 当てはまり具合の基準: CC |
| 得られたモデル |  PDB-8peg: |
 ムービー
ムービー コントローラー
コントローラー






















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)