+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 10-mer ring of human metapneumovirus (HMPV) N-RNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nucleoprotein / Virus / Nucleocapsid / RNA-binding / HMPV / Pneumoviridae / Mononegavirales / VIRAL PROTEIN | |||||||||
| Function / homology | Pneumovirus nucleocapsid protein / Pneumovirus nucleocapsid protein / helical viral capsid / viral nucleocapsid / host cell cytoplasm / ribonucleoprotein complex / RNA binding / Nucleoprotein Function and homology information Function and homology information | |||||||||
| Biological species |  Human metapneumovirus (strain CAN97-83) / Human metapneumovirus (strain CAN97-83) /  | |||||||||
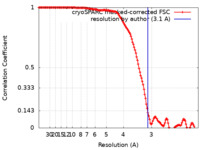
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Whitehead JD / Decool H / Leyrat C / Carrique L / Fix J / Eleouet JF / Galloux M / Renner M | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus. Authors: Jack D Whitehead / Hortense Decool / Cédric Leyrat / Loic Carrique / Jenna Fix / Jean-François Eléouët / Marie Galloux / Max Renner /    Abstract: Human metapneumovirus (HMPV) is a major cause of respiratory illness in young children. The HMPV polymerase (L) binds an obligate cofactor, the phosphoprotein (P). During replication and ...Human metapneumovirus (HMPV) is a major cause of respiratory illness in young children. The HMPV polymerase (L) binds an obligate cofactor, the phosphoprotein (P). During replication and transcription, the L/P complex traverses the viral RNA genome, which is encapsidated within nucleoproteins (N). An essential interaction between N and a C-terminal region of P tethers the L/P polymerase to the template. This N-P interaction is also involved in the formation of cytoplasmic viral factories in infected cells, called inclusion bodies. To define how the polymerase component P recognizes N-encapsidated RNA (N-RNA) we employed cryogenic electron microscopy (cryo-EM) and molecular dynamics simulations, coupled to activity assays and imaging of inclusion bodies in cells. We report a 2.9 Å resolution structure of a triple-complex between multimeric N, bound to both RNA and the C-terminal region of P. Furthermore, we also present cryo-EM structures of assembled N in different oligomeric states, highlighting the plasticity of N. Combined with our functional assays, these structural data delineate in molecular detail how P attaches to N-RNA whilst retaining substantial conformational dynamics. Moreover, the N-RNA-P triple complex structure provides a molecular blueprint for the design of therapeutics to potentially disrupt the attachment of L/P to its template. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17613.map.gz emd_17613.map.gz | 230.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17613-v30.xml emd-17613-v30.xml emd-17613.xml emd-17613.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17613_fsc.xml emd_17613_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_17613.png emd_17613.png | 122.6 KB | ||
| Filedesc metadata |  emd-17613.cif.gz emd-17613.cif.gz | 5.9 KB | ||
| Others |  emd_17613_half_map_1.map.gz emd_17613_half_map_1.map.gz emd_17613_half_map_2.map.gz emd_17613_half_map_2.map.gz | 226.4 MB 226.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17613 http://ftp.pdbj.org/pub/emdb/structures/EMD-17613 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17613 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17613 | HTTPS FTP |
-Related structure data
| Related structure data |  8pdlMC  8pdmC  8pdnC  8pdoC  8pdpC  8pdqC  8pdrC  8pdsC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17613.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17613.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_17613_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17613_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human metapneumovirus N-RNA
| Entire | Name: Human metapneumovirus N-RNA |
|---|---|
| Components |
|
-Supramolecule #1: Human metapneumovirus N-RNA
| Supramolecule | Name: Human metapneumovirus N-RNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 460 KDa |
-Supramolecule #2: Nucleoprotein
| Supramolecule | Name: Nucleoprotein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus (strain CAN97-83) / Strain: CAN97-83 Human metapneumovirus (strain CAN97-83) / Strain: CAN97-83 |
-Supramolecule #3: RNA
| Supramolecule | Name: RNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Nucleoprotein
| Macromolecule | Name: Nucleoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus (strain CAN97-83) / Strain: CAN97-83 Human metapneumovirus (strain CAN97-83) / Strain: CAN97-83 |
| Molecular weight | Theoretical: 43.57652 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSLQGIHLSD LSYKHAILKE SQYTIKRDVG TTTAVTPSSL QQEITLLCGE ILYAKHADYK YAAEIGIQYI STALGSERVQ QILRNSGSE VQVVLTRTYS LGKIKNNKGE DLQMLDIHGV EKSWVEEIDK EARKTMATLL KESSGNIPQN QRPSAPDTPI I LLCVGALI ...String: MSLQGIHLSD LSYKHAILKE SQYTIKRDVG TTTAVTPSSL QQEITLLCGE ILYAKHADYK YAAEIGIQYI STALGSERVQ QILRNSGSE VQVVLTRTYS LGKIKNNKGE DLQMLDIHGV EKSWVEEIDK EARKTMATLL KESSGNIPQN QRPSAPDTPI I LLCVGALI FTKLASTIEV GLETTVRRAN RVLSDALKRY PRMDIPKIAR SFYDLFEQKV YHRSLFIEYG KALGSSSTGS KA ESLFVNI FMQAYGAGQT MLRWGVIARS SNNIMLGHVS VQAELKQVTE VYDLVREMGP ESGLLHLRQS PKAGLLSLAN CPN FASVVL GNASGLGIIG MYRGRVPNTE LFSAAESYAK SLKESNKINF SSLGLTDEEK EAAEHFLNVS DDSQNDYE UniProtKB: Nucleoprotein |
-Macromolecule #2: RNA
| Macromolecule | Name: RNA / type: rna / ID: 2 Details: E.coli RNA, co-purified with N. Due to mixed sequence, modelled as poly-C Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.091315 KDa |
| Sequence | String: CCCCCCC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 25 mM Tris-HCl, pH 8.0, 250 mM NaCl |
| Grid | Model: Quantifoil R2/1 / Material: COPPER |
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 44.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)