+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ligand-free SpSLC9C1 in detergent, asymmetric class | |||||||||||||||
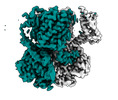
 Map data Map data | deepEMhancer sharpened map of ligand-free SpSLC9C1 in detergent, asymmetric class | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | SLC9 / NHE / sperm-specific / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsperm head / potassium:proton antiporter activity / sodium:proton antiporter activity / sodium ion import across plasma membrane / cGMP binding / sperm flagellum / single fertilization / cAMP binding / potassium ion transmembrane transport / regulation of intracellular pH ...sperm head / potassium:proton antiporter activity / sodium:proton antiporter activity / sodium ion import across plasma membrane / cGMP binding / sperm flagellum / single fertilization / cAMP binding / potassium ion transmembrane transport / regulation of intracellular pH / protein homodimerization activity / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
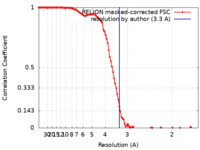
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||||||||
 Authors Authors | Kalienkova V / Peter M / Rheinberger J / Paulino C | |||||||||||||||
| Funding support |  Netherlands, Netherlands,  Switzerland, 4 items Switzerland, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Structures of a sperm-specific solute carrier gated by voltage and cAMP. Authors: Valeria Kalienkova / Martin F Peter / Jan Rheinberger / Cristina Paulino /    Abstract: The newly characterized sperm-specific Na/H exchanger stands out by its unique tripartite domain composition. It unites a classical solute carrier unit with regulatory domains usually found in ion ...The newly characterized sperm-specific Na/H exchanger stands out by its unique tripartite domain composition. It unites a classical solute carrier unit with regulatory domains usually found in ion channels, namely, a voltage-sensing domain and a cyclic-nucleotide binding domain, which makes it a mechanistic chimera and a secondary-active transporter activated strictly by membrane voltage. Our structures of the sea urchin SpSLC9C1 in the absence and presence of ligands reveal the overall domain arrangement and new structural coupling elements. They allow us to propose a gating model, where movements in the voltage sensor indirectly cause the release of the exchanging unit from a locked state through long-distance allosteric effects transmitted by the newly characterized coupling helices. We further propose that modulation by its ligand cyclic AMP occurs by means of disruption of the cytosolic dimer interface, which lowers the energy barrier for S4 movements in the voltage-sensing domain. As SLC9C1 members have been shown to be essential for male fertility, including in mammals, our structure represents a potential new platform for the development of new on-demand contraceptives. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17603.map.gz emd_17603.map.gz | 91.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17603-v30.xml emd-17603-v30.xml emd-17603.xml emd-17603.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17603_fsc.xml emd_17603_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_17603.png emd_17603.png | 98.3 KB | ||
| Masks |  emd_17603_msk_1.map emd_17603_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17603.cif.gz emd-17603.cif.gz | 6.1 KB | ||
| Others |  emd_17603_additional_1.map.gz emd_17603_additional_1.map.gz emd_17603_half_map_1.map.gz emd_17603_half_map_1.map.gz emd_17603_half_map_2.map.gz emd_17603_half_map_2.map.gz | 79.5 MB 80.2 MB 80.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17603 http://ftp.pdbj.org/pub/emdb/structures/EMD-17603 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17603 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17603 | HTTPS FTP |
-Validation report
| Summary document |  emd_17603_validation.pdf.gz emd_17603_validation.pdf.gz | 697.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17603_full_validation.pdf.gz emd_17603_full_validation.pdf.gz | 697.1 KB | Display | |
| Data in XML |  emd_17603_validation.xml.gz emd_17603_validation.xml.gz | 17.7 KB | Display | |
| Data in CIF |  emd_17603_validation.cif.gz emd_17603_validation.cif.gz | 22.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17603 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17603 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17603 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17603 | HTTPS FTP |
-Related structure data
| Related structure data |  8pczC  8pd2C  8pd3C  8pd5C  8pd7C  8pd8C  8pd9C  8pduC  8pdvC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17603.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17603.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepEMhancer sharpened map of ligand-free SpSLC9C1 in detergent, asymmetric class | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.836 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17603_msk_1.map emd_17603_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: refined map of ligand-free SpSLC9C1 in detergent, asymmetric class
| File | emd_17603_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | refined map of ligand-free SpSLC9C1 in detergent, asymmetric class | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17603_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17603_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ligand-free SpSLC9C1 in detergent, asymmetric class
| Entire | Name: Ligand-free SpSLC9C1 in detergent, asymmetric class |
|---|---|
| Components |
|
-Supramolecule #1: Ligand-free SpSLC9C1 in detergent, asymmetric class
| Supramolecule | Name: Ligand-free SpSLC9C1 in detergent, asymmetric class / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: SpSLC9C1
| Macromolecule | Name: SpSLC9C1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MSKKRVVKLR ELVPAVAALA VAVLIQSATG SSGGSGHTPT TQATHADDHD LTTHNGTEEH DDGHDDGHDD LHAHAPKVIV FISGSCLFGA ISRSLFKKLP IPYTVVLLIL GAILGVVASN VPLVEEHTRD VAHMDPHVLL QIFLPVLIFE SAFAMDVHTF MRSFSQVCIL ...String: MSKKRVVKLR ELVPAVAALA VAVLIQSATG SSGGSGHTPT TQATHADDHD LTTHNGTEEH DDGHDDGHDD LHAHAPKVIV FISGSCLFGA ISRSLFKKLP IPYTVVLLIL GAILGVVASN VPLVEEHTRD VAHMDPHVLL QIFLPVLIFE SAFAMDVHTF MRSFSQVCIL ALFGLVVASV LTAVLAMNLF NYNWNFSEAM MFGAIMSATD PVAVVALLKD LGASKQLGTI IEGESLLNDG CAIVIFNVFM KMVFFPQLTS TVGQNVLYFL QVAVAGPLWG YAVAKVTVFF LSHIFNDALV EITITLAATY LTYYIGDIWL EVSGVLAVVV LGLIVNAEKT SISPEVEVFL HRFWEMLAYL ANTLIFMMVG VVVTQKALVA VDKMDWFYLI ILYLAITIIR GMVISLFSPI LSRIGYGLTW RNAVIMTWGG LRGAVGLALA LVVENLAGND VIGSKFLFHT AGIVVLTLVI NATTIQTLLR ILGMSDISIP KRLAMAGAVR RIHEGQNRTL NMLKSDRFLA DADWDIATAA CEISDPYSAL SDDENAPADE LTLGERKSVC PGCKAMVPNE PSPREFADMM EEARLRMLKA EKISYWKQFE HGMLAREALR LLVQHAEVAA DEKDQFILVD DLKKSWQIKG IYPWLKRKLE DLISEKKIAA IPMPKYKLGK LMYKICHHMA FEVTINIAIV LNIVPIIMEF VVQDKMASVS TMAAPGSTVS SEPSSLQKIE DALRISNYVF FVIYAIEAIV KILGLGRHYI VSHWNKFDAF ILVVALVDII IAETLLKGSI TINLSSIKVV KLFRLLRGLR MLRLTKALIP KLILVVNGKI NNQLSLGYDV GKGYIIGEEE VGKIIDRMVD NKKILRELKH ISETGRLQVV KELGLLQREH PGIAVSVKTR QAIRTILNHS RETIHELQGA GLLDEMEAHK LELTVEIKMK RLMNAPSSIP PPPPENLLKN VSWLAGDMKL IDFIKARASL LHFDYGEVIV REGDESDGLF LIVSGLVKLY GKSAFLDHDN PPVTAGSEEN EVFEDYLTVG NVIGEMGVLT KKPRNATVTC ETTVQVYFIT AEDMNIAIDT FTLYPSLEYR LWRVVAIRIA TPLIMEQMAF QGWTQEKVKL HLERGYLVDL AESHFQFNID ATLEDVILIN GTAYNAHTRE EIRSPCLISR TVHKLTFQYT ATEEPRLFVV RNAEYNGPIL DGRLDVDSKR SLISITEISS NMCLKHAAEL RQKNSKVMLS RKSSGAAAKE EEDCIPNTSD VEQAAGVSPS VPTKTTPKPK SFLPSLGLSM SKERVNGEAV EESPVKTKQG EETPETEEGA APRVNVALEV LFQ UniProtKB: Sperm-specific sodium:proton exchanger |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.93 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| ||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 288.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 2 / Number real images: 7090 / Average exposure time: 2.31 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.2 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)