+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ex vivo L-ENA type pili from Bacillus paranthracis endospores | |||||||||
 Map data Map data | Ex vivo L-ENA map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Endospore / Pili / Protein / Fiber / Spore / Appendage / Bacillus paranthracis / PROTEIN FIBRIL | |||||||||
| Function / homology | :  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
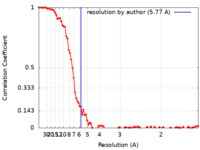
| Method | helical reconstruction / cryo EM / Resolution: 5.77 Å | |||||||||
 Authors Authors | Sleutel M / Remaut H | |||||||||
| Funding support |  Belgium, 1 items Belgium, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Helical ultrastructure of the L-ENA spore aggregation factor of a Bacillus paranthracis foodborne outbreak strain. Authors: Mike Sleutel / Ephrem Debebe Zegeye / Ann-Katrin Llarena / Brajabandhu Pradhan / Marcus Fislage / Kristin O'Sullivan / Nani Van Gerven / Marina Aspholm / Han Remaut /   Abstract: In pathogenic Bacillota, spores can form an infectious particle and can take up a central role in the environmental persistence and dissemination of disease. A poorly understood aspect of spore- ...In pathogenic Bacillota, spores can form an infectious particle and can take up a central role in the environmental persistence and dissemination of disease. A poorly understood aspect of spore-mediated infection is the fibrous structures or 'endospore appendages' (ENAs) that have been seen to decorate the spores of pathogenic Bacilli and Clostridia. Current methodological approaches are opening a window on these long enigmatic structures. Using cryoID, Alphafold modelling and genetic approaches we identify a sub-class of robust ENAs in a Bacillus paranthracis foodborne outbreak strain. We demonstrate that L-ENA are encoded by a rare three-gene cluster (ena3) that contains all components for the self-assembly of ladder-like protein nanofibers of stacked heptameric rings, their anchoring to the exosporium, and their termination in a trimeric 'ruffle' made of a complement C1Q-like BclA paralogue. The role of ENA fibers in spore-spore interaction and the distribution of L-ENA operon as mobile genetic elements in B. cereus s.l. strains suggest that L-ENA fibers may increase the survival, spread and virulence of these strains. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17579.map.gz emd_17579.map.gz | 31.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17579-v30.xml emd-17579-v30.xml emd-17579.xml emd-17579.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17579_fsc.xml emd_17579_fsc.xml | 11.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_17579.png emd_17579.png | 62.1 KB | ||
| Masks |  emd_17579_msk_1.map emd_17579_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17579.cif.gz emd-17579.cif.gz | 4.7 KB | ||
| Others |  emd_17579_half_map_1.map.gz emd_17579_half_map_1.map.gz emd_17579_half_map_2.map.gz emd_17579_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17579 http://ftp.pdbj.org/pub/emdb/structures/EMD-17579 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17579 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17579 | HTTPS FTP |
-Related structure data
| Related structure data |  8pdzC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17579.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17579.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ex vivo L-ENA map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.766 Å | ||||||||||||||||||||||||||||||||||||
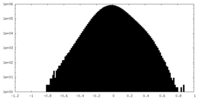
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17579_msk_1.map emd_17579_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
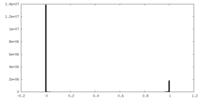
| Density Histograms |
-Half map: Ex vivo L-ENA half map A
| File | emd_17579_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ex vivo L-ENA half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
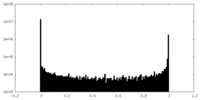
| Density Histograms |
-Half map: Ex vivo L-ENA half map B
| File | emd_17579_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ex vivo L-ENA half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
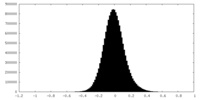
| Density Histograms |
- Sample components
Sample components
-Entire : L-type endospore appendage
| Entire | Name: L-type endospore appendage |
|---|---|
| Components |
|
-Supramolecule #1: L-type endospore appendage
| Supramolecule | Name: L-type endospore appendage / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.03 kDa/nm |
-Macromolecule #1: Ena3A
| Macromolecule | Name: Ena3A / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MAQIGNCCTE QLCCVNDAVC CTIILDDTGG TALPIWDDAT TFVINGTIMV ENNGTVGVGP TAALTVNGTA VGGFVVAPGE CRSITMNDIN SIAIVGAGTG TSSVKISFSI NYKF UniProtKB: UNIPROTKB: PGS39_28750 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Component - Formula: miliQ |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number real images: 8528 / Average electron dose: 62.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 60000 |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)