[English] 日本語
 Yorodumi
Yorodumi- EMDB-17459: S. cerevisiae consensus-sCMGE on ssDNA after DNA replication init... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | S. cerevisiae consensus-sCMGE on ssDNA after DNA replication initiation | |||||||||||||||
 Map data Map data | EM map of consensus-sCMGE | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Saccharomyces cerevisiae / helicase / CMGE / initiation of DNA replication / DNA / DNA unwinding / REPLICATION | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationDNA-templated DNA replication maintenance of fidelity / gene conversion / Unwinding of DNA / DNA replication initiation / epsilon DNA polymerase complex / MCM core complex / Assembly of the pre-replicative complex / Switching of origins to a post-replicative state / DNA strand elongation involved in mitotic DNA replication / GINS complex ...DNA-templated DNA replication maintenance of fidelity / gene conversion / Unwinding of DNA / DNA replication initiation / epsilon DNA polymerase complex / MCM core complex / Assembly of the pre-replicative complex / Switching of origins to a post-replicative state / DNA strand elongation involved in mitotic DNA replication / GINS complex / MCM complex binding / mitotic DNA replication preinitiation complex assembly / nuclear DNA replication / premeiotic DNA replication / replication fork protection complex / pre-replicative complex assembly involved in nuclear cell cycle DNA replication / SUMO binding / nucleotide-excision repair, DNA gap filling / Activation of the pre-replicative complex / mitotic DNA replication / CMG complex / nuclear pre-replicative complex / DNA replication proofreading / Termination of translesion DNA synthesis / Activation of ATR in response to replication stress / DNA replication preinitiation complex / single-stranded DNA 3'-5' DNA exonuclease activity / MCM complex / mitotic DNA replication checkpoint signaling / double-strand break repair via break-induced replication / mitotic DNA replication initiation / single-stranded DNA helicase activity / mitotic intra-S DNA damage checkpoint signaling / silent mating-type cassette heterochromatin formation / Hydrolases; Acting on ester bonds; Exodeoxyribonucleases producing 5'-phosphomonoesters / regulation of DNA-templated DNA replication initiation / mitotic sister chromatid cohesion / DNA strand elongation involved in DNA replication / leading strand elongation / nuclear replication fork / DNA replication origin binding / Dual incision in TC-NER / DNA replication initiation / error-prone translesion synthesis / subtelomeric heterochromatin formation / base-excision repair, gap-filling / DNA helicase activity / replication fork / transcription elongation by RNA polymerase II / helicase activity / base-excision repair / double-strand break repair via nonhomologous end joining / DNA-templated DNA replication / heterochromatin formation / double-strand break repair / mitotic cell cycle / single-stranded DNA binding / 4 iron, 4 sulfur cluster binding / double-stranded DNA binding / DNA helicase / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / chromosome, telomeric region / DNA replication / nucleotide binding / mRNA binding / DNA damage response / chromatin binding / ATP hydrolysis activity / DNA binding / zinc ion binding / nucleoplasm / ATP binding / nucleus / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
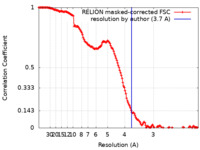
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||||||||
 Authors Authors | Henrikus SS / Willhoft O | |||||||||||||||
| Funding support |  United Kingdom, European Union, United Kingdom, European Union,  France, 4 items France, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Unwinding of a eukaryotic origin of replication visualized by cryo-EM. Authors: Sarah S Henrikus / Marta H Gross / Oliver Willhoft / Thomas Pühringer / Jacob S Lewis / Allison W McClure / Julia F Greiwe / Giacomo Palm / Andrea Nans / John F X Diffley / Alessandro Costa /    Abstract: To prevent detrimental chromosome re-replication, DNA loading of a double hexamer of the minichromosome maintenance (MCM) replicative helicase is temporally separated from DNA unwinding. Upon S-phase ...To prevent detrimental chromosome re-replication, DNA loading of a double hexamer of the minichromosome maintenance (MCM) replicative helicase is temporally separated from DNA unwinding. Upon S-phase transition in yeast, DNA unwinding is achieved in two steps: limited opening of the double helix and topological separation of the two DNA strands. First, Cdc45, GINS and Polε engage MCM to assemble a double CMGE with two partially separated hexamers that nucleate DNA melting. In the second step, triggered by Mcm10, two CMGEs separate completely, eject the lagging-strand template and cross paths. To understand Mcm10 during helicase activation, we used biochemical reconstitution with cryogenic electron microscopy. We found that Mcm10 splits the double CMGE by engaging the N-terminal homo-dimerization face of MCM. To eject the lagging strand, DNA unwinding is started from the N-terminal side of MCM while the hexamer channel becomes too narrow to harbor duplex DNA. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17459.map.gz emd_17459.map.gz | 263.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17459-v30.xml emd-17459-v30.xml emd-17459.xml emd-17459.xml | 43.7 KB 43.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17459_fsc.xml emd_17459_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_17459.png emd_17459.png | 166.7 KB | ||
| Filedesc metadata |  emd-17459.cif.gz emd-17459.cif.gz | 13.3 KB | ||
| Others |  emd_17459_half_map_1.map.gz emd_17459_half_map_1.map.gz emd_17459_half_map_2.map.gz emd_17459_half_map_2.map.gz | 225.2 MB 225.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17459 http://ftp.pdbj.org/pub/emdb/structures/EMD-17459 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17459 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17459 | HTTPS FTP |
-Validation report
| Summary document |  emd_17459_validation.pdf.gz emd_17459_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17459_full_validation.pdf.gz emd_17459_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_17459_validation.xml.gz emd_17459_validation.xml.gz | 22.3 KB | Display | |
| Data in CIF |  emd_17459_validation.cif.gz emd_17459_validation.cif.gz | 29.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17459 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17459 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17459 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17459 | HTTPS FTP |
-Related structure data
| Related structure data |  8p63MC  8p5eC  8p62C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17459.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17459.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map of consensus-sCMGE | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
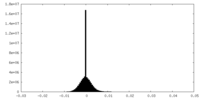
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: EM half map 1 of consensus-sCMGE
| File | emd_17459_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half map 1 of consensus-sCMGE | ||||||||||||
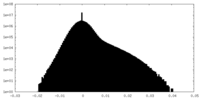
| Projections & Slices |
| ||||||||||||
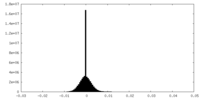
| Density Histograms |
-Half map: EM half map 2 of consensus-sCMGE
| File | emd_17459_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half map 2 of consensus-sCMGE | ||||||||||||
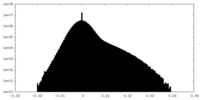
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Complex of Mcm2-7, Cdc45, GINS (Psf1-3, Sld5) DNA polymerase epsi...
+Supramolecule #1: Complex of Mcm2-7, Cdc45, GINS (Psf1-3, Sld5) DNA polymerase epsi...
+Supramolecule #2: DNA replication complex GINS containing Psf1, Psf2, Psf3 and Sld5
+Supramolecule #3: DNA
+Supramolecule #4: DNA replication licensing factors MCM2-7
+Supramolecule #5: DNA polymerase epsilon contains catalytic subunit A and non-catal...
+Supramolecule #6: Cdc45
+Macromolecule #1: DNA replication licensing factor MCM2
+Macromolecule #2: DNA replication licensing factor MCM3
+Macromolecule #3: DNA replication licensing factor MCM4
+Macromolecule #4: Minichromosome maintenance protein 5
+Macromolecule #5: DNA replication licensing factor MCM6
+Macromolecule #6: DNA replication licensing factor MCM7
+Macromolecule #8: DNA replication complex GINS protein PSF3
+Macromolecule #9: DNA replication complex GINS protein SLD5
+Macromolecule #10: Cell division control protein 45
+Macromolecule #11: DNA polymerase epsilon subunit B
+Macromolecule #12: DNA polymerase epsilon catalytic subunit A
+Macromolecule #13: DNA replication complex GINS protein PSF1
+Macromolecule #14: DNA replication complex GINS protein PSF2
+Macromolecule #7: DNA (9-MER)
+Macromolecule #15: ADENOSINE-5'-TRIPHOSPHATE
+Macromolecule #16: ZINC ION
+Macromolecule #17: MAGNESIUM ION
+Macromolecule #18: ADENOSINE-5'-DIPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.67 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.3000000000000003 µm / Nominal defocus min: 1.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)