[English] 日本語
 Yorodumi
Yorodumi- EMDB-17352: Structure of hantaan orthohantavirus (HTNV) polymerase - Apo core... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of hantaan orthohantavirus (HTNV) polymerase - Apo core endonuclease | ||||||||||||
 Map data Map data | Global sharpening | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | RNA-dependent RNA polymerase / HTNV / Hantaan orthohantavirus / Bunyavirales / Hantaviridae. / VIRAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationRNA-templated viral transcription / negative stranded viral RNA replication / cap snatching / endonuclease activity / Hydrolases; Acting on ester bonds / host cell perinuclear region of cytoplasm / RNA-directed RNA polymerase / nucleotide binding / RNA-directed RNA polymerase activity / DNA-templated transcription / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Hantaan orthohantavirus Hantaan orthohantavirus | ||||||||||||
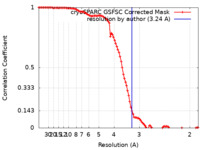
| Method | single particle reconstruction / cryo EM / Resolution: 3.24 Å | ||||||||||||
 Authors Authors | Keown JR / Carrique L / Grimes JM | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2024 Journal: PLoS Pathog / Year: 2024Title: Structural characterization of the full-length Hantaan virus polymerase. Authors: Jeremy R Keown / Loïc Carrique / Benjamin E Nilsson-Payant / Ervin Fodor / Jonathan M Grimes /    Abstract: Hantaviridae are a family of segmented negative-sense RNA viruses that contain important human and animal pathogens. Hantaviridae contain a viral RNA-dependent RNA polymerase that replicates and ...Hantaviridae are a family of segmented negative-sense RNA viruses that contain important human and animal pathogens. Hantaviridae contain a viral RNA-dependent RNA polymerase that replicates and transcribes the viral genome. Here we establish the expression and purification of the polymerase from the Old World Hantaan virus and characterise the structure using Cryo-EM. We determine a series of structures at resolutions between 2.7 and 3.3 Å of RNA free polymerase comprising the core, core and endonuclease, and a full-length polymerase. The full-length polymerase structure depicts the location of the cap binding and C-terminal domains which are arranged in a conformation that is incompatible with transcription and in a novel conformation not observed in previous conformations of cap-snatching viral polymerases. We further describe structures with 5' vRNA promoter in the presence and absence of a nucleotide triphosphate. The nucleotide bound structure mimics a replication pre-initiation complex and the nucleotide stabilises the motif E in a conformation distinct from those previously observed. We observe motif E in four distinct conformations including β-sheet, two helical arrangements, and nucleotide primed arrangement. The insights gained here guide future mechanistic studies of both the transcription and replication activities of the hantavirus polymerase and for the development of therapeutic targets. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17352.map.gz emd_17352.map.gz | 97.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17352-v30.xml emd-17352-v30.xml emd-17352.xml emd-17352.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17352_fsc.xml emd_17352_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_17352.png emd_17352.png | 100.3 KB | ||
| Masks |  emd_17352_msk_1.map emd_17352_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17352.cif.gz emd-17352.cif.gz | 7.4 KB | ||
| Others |  emd_17352_additional_1.map.gz emd_17352_additional_1.map.gz emd_17352_half_map_1.map.gz emd_17352_half_map_1.map.gz emd_17352_half_map_2.map.gz emd_17352_half_map_2.map.gz | 95.5 MB 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17352 http://ftp.pdbj.org/pub/emdb/structures/EMD-17352 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17352 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17352 | HTTPS FTP |
-Related structure data
| Related structure data |  8p1kMC  8p1jC  8p1lC  8p1mC  8p1nC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17352.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17352.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Global sharpening | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.932 Å | ||||||||||||||||||||||||||||||||||||
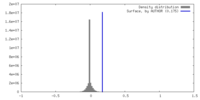
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17352_msk_1.map emd_17352_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
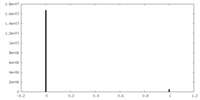
| Density Histograms |
-Additional map: deepEMhancer sharpening
| File | emd_17352_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepEMhancer sharpening | ||||||||||||
| Projections & Slices |
| ||||||||||||
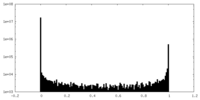
| Density Histograms |
-Half map: #1
| File | emd_17352_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
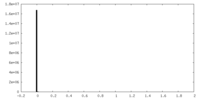
| Density Histograms |
-Half map: #2
| File | emd_17352_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RNA-dependent RNA polymerase of Hantaan orthohantavirus (HTNV).
| Entire | Name: RNA-dependent RNA polymerase of Hantaan orthohantavirus (HTNV). |
|---|---|
| Components |
|
-Supramolecule #1: RNA-dependent RNA polymerase of Hantaan orthohantavirus (HTNV).
| Supramolecule | Name: RNA-dependent RNA polymerase of Hantaan orthohantavirus (HTNV). type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Hantaan orthohantavirus Hantaan orthohantavirus |
| Molecular weight | Theoretical: 247 KDa |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Hantaan orthohantavirus Hantaan orthohantavirus |
| Molecular weight | Theoretical: 251.806016 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MWSHPQFEKG GGSGGGSGGS SAWSHPQFEK HHHHHHHHEN LYFQGMDKYR EIHNKLKEFS PGTLTAVECI DYLDRLYAVR HDIVDQMIK HDWSDNKDSE EAIGKVLLFA GVPSNIITAL EKKIIPNHPT GKSLKAFFKM TPANYKISGT TIEFVEVTVT A DVDKGIRE ...String: MWSHPQFEKG GGSGGGSGGS SAWSHPQFEK HHHHHHHHEN LYFQGMDKYR EIHNKLKEFS PGTLTAVECI DYLDRLYAVR HDIVDQMIK HDWSDNKDSE EAIGKVLLFA GVPSNIITAL EKKIIPNHPT GKSLKAFFKM TPANYKISGT TIEFVEVTVT A DVDKGIRE KKLKYEAGLT YIEQELHKFF LKGEIPQPYK ITFNVVAVRT DGSNITTQWP SRRNDGVVQY MRLVQAEISY VR EHLIKTE ERAALEAMFN LKFNISTHKS QPYYIPDYKG MEPIGANIED LVDYSKDWLS RARNFSFFEV KGTAVFECFN SNE ANHCQR YPMSRKPRNF LLIQCSLITS YKPATTLSDQ IDSRRACSYI LNLIPDTPAS YLIHDMAYRY INLTREDMIN YYAP RIQFK QTQNVREPGT FKLTSSMLRA ESKAMLDLLN NHKSGEKHGA QIESLNIASH IVQSESVSLI TKILSDLELN ITEPS TQEY STTKHTYVDT VLDKFFQNET QKYLIDVLKK TTAWHIGHLI RDITESLIAH SGLKRSKYWS LHSYNNGNVI LFILPS KSL EVAGSFIRFI TVFRIGPGLV DKDNLDTILI DGDSQWGVSK VMSIDLNRLL ALNIAFEKAL IATATWFQYY TEDQGQF PL QYAIRSVFAN HFLLAICQKM KLCAIFDNLR YLIPAVTSLY SGFPSLIEKL FERPFKSSLE VYIYYNIKSL LVALAQNN K ARFYSKVKLL GLTVDQSTVG ASGVYPSFMS RIVYKHYRSL ISEVTTCFFL FEKGLHGNMN EEAKIHLETV EWALKFREK EEKYGESLVE NGYMMWELRA NAELAEQQLY CQDAIELAAI ELNKVLATKS SVVANSILSK NWEEPYFSQT RNISLKGMSG QVQEDGHLS SSVTIIEAIR YLSNSRHNPS LLKLYEETRE QKAMARIVRK YQRTEADRGF FITTLPTRCR LEIIEDYYDA I AKNISEEY ISYGGEKKIL AIQGALEKAL RWASGESFIE LSNHKFIRMK RKLMYVSADA TKWSPGDNSA KFRRFTSMLH NG LPNNKLK NCVIDALKQV YKTDFFMSRK LRNYIDSMES LDPHIKQFLD FFPDGHHGEV KGNWLQGNLN KCSSLFGVAM SLL FKQVWT NLFPELDCFF EFAHHSDDAL FIYGYLEPVD DGTDWFLFVS QQIQAGHLHW FSVNTEMWKS MFNLHEHILL LGSI KISPK KTTVSPTNAE FLSTFFEGCA VSIPFVKILL GSLSDLPGLG YFDDLAAAQS RCVKALDLGA SPQVAQLAVA LCTSK VERL YGTAPGMVNH PAAYLQVKHT DTPIPLGGNG AMSIMELATA GIGMSDKNLL KRALLGYSHK RQKSMLYILG LFKFLM KLS DETFQHERLG QFSFIGKVQW KIFTPKSEFE FADMYTSKFL ELWSSQHVTY DYIIPKGRDN LLIYLVRKLN DPSIVTA MT MQSPLQLRFR MQAKQHMKVC RLDGEWVTFR EVLAAANSFA ENYSATSQDM DLFQTLTSCT FSKEYAWKDF LNGIHCDV I PTKQVQRAKV ARTFTVREKD QIIQNSIPAV IGYKFAVTVE EMSDVLDTAK FPDSLSVDLK TMKDGVYREL GLDISLPDV MKRIAPMLYK SSKSRVVIVQ GNVEGTAEAI CRYWLKSMSL VKTIRVKPHK EVLQAVSIFN RKEDIGQQKD LAALKLCIEV WRWCKANSA PYRDWFQALW FEDKTFSEWL DRFCRVGVPP IDPEIQCAAL MIADIKGDYS VLQLQANRRA YSGKQYDAYC V QTYNEVTK LYEGDLRVTF NFGLDCARLE IFWDKKAYIL ETSITQKHVL KIMMDEVSKE LIKCGMRFNT EQVQGVRHMV LF KTESGFE WGKPNIPCIV YKNCVLRTSL RTTQAINHKF MITIKDDGLR AIAQHDEDSP RFLLAHAFHT IRDIRYQAVD AVS NVWFIH KGVKLYLNPI ISSGLLENFM KNLPAAIPPA AYSLIMNRAK ISVDLFMFND LLKLINPRNT LDLSGLETTG DEFS TVSSM SSRLWSEEMS LVDDDEELDD EFTIDLQDVD FENIDIEADI EHFLQDESSY TGDLLISTEE TESKKMRGIV KILEP VRLI KSWVSRGLSI EKVYSPVNII LMSRYISKTF NLSTKQVSLL DPYDLTELES IVRGWGECVI DQFESLDREA QNMVVN KGI CPEDVIPDSL FSFRHTMVLL RRLFPQDSIS SFY UniProtKB: RNA-directed RNA polymerase L |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.4000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Cross-correlation coefficient |
| Output model |  PDB-8p1k: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)