[English] 日本語
 Yorodumi
Yorodumi- EMDB-17298: Cryo-EM structure of Phthaloyl-CoA decarboxylase (Pcd) bound with... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Phthaloyl-CoA decarboxylase (Pcd) bound with substrate analog/inhibitor, 2-CN-benzoyl-CoA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | prFMN / plastic degradation / Phthalates / anaerobic / light sensitive / Co-enzyme A / decarboxylase / cyano-benzoyl CoA / inhibitor bound / substrate analog / FLAVOPROTEIN | |||||||||
| Biological species |  Thauera chlorobenzoica (bacteria) Thauera chlorobenzoica (bacteria) | |||||||||
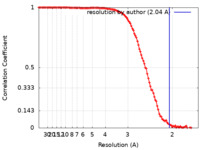
| Method | single particle reconstruction / cryo EM / Resolution: 2.04 Å | |||||||||
 Authors Authors | Kayastha K / Ermler U | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of Phthaloyl-CoA decarboxylase (Pcd) bound with substrate analog/inhibitor, 2-CN-benzoyl-CoA Authors: Kayastha K / Ermler U | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17298.map.gz emd_17298.map.gz | 97.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17298-v30.xml emd-17298-v30.xml emd-17298.xml emd-17298.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17298_fsc.xml emd_17298_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_17298.png emd_17298.png | 163.6 KB | ||
| Filedesc metadata |  emd-17298.cif.gz emd-17298.cif.gz | 6.2 KB | ||
| Others |  emd_17298_half_map_1.map.gz emd_17298_half_map_1.map.gz emd_17298_half_map_2.map.gz emd_17298_half_map_2.map.gz | 98.1 MB 98.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17298 http://ftp.pdbj.org/pub/emdb/structures/EMD-17298 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17298 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17298 | HTTPS FTP |
-Validation report
| Summary document |  emd_17298_validation.pdf.gz emd_17298_validation.pdf.gz | 743 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17298_full_validation.pdf.gz emd_17298_full_validation.pdf.gz | 742.5 KB | Display | |
| Data in XML |  emd_17298_validation.xml.gz emd_17298_validation.xml.gz | 18.1 KB | Display | |
| Data in CIF |  emd_17298_validation.cif.gz emd_17298_validation.cif.gz | 24.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17298 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17298 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17298 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17298 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17298.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17298.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.837 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_17298_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
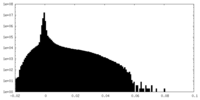
| Projections & Slices |
| ||||||||||||
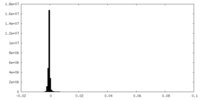
| Density Histograms |
-Half map: #2
| File | emd_17298_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
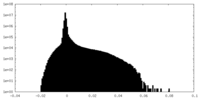
| Projections & Slices |
| ||||||||||||
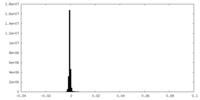
| Density Histograms |
- Sample components
Sample components
-Entire : Phthaloyl-CoA decarboxylase (Pcd) bound with 2-CN-benzoyl-CoA
| Entire | Name: Phthaloyl-CoA decarboxylase (Pcd) bound with 2-CN-benzoyl-CoA |
|---|---|
| Components |
|
-Supramolecule #1: Phthaloyl-CoA decarboxylase (Pcd) bound with 2-CN-benzoyl-CoA
| Supramolecule | Name: Phthaloyl-CoA decarboxylase (Pcd) bound with 2-CN-benzoyl-CoA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Thauera chlorobenzoica (bacteria) Thauera chlorobenzoica (bacteria) |
| Molecular weight | Theoretical: 360 KDa |
-Macromolecule #1: Phthaloyl-CoA decarboxylase (Pcd)
| Macromolecule | Name: Phthaloyl-CoA decarboxylase (Pcd) / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thauera chlorobenzoica (bacteria) Thauera chlorobenzoica (bacteria) |
| Molecular weight | Theoretical: 57.682492 KDa |
| Sequence | String: ERVGEKDLRA ALEWFRSKGY LVETNKEVNP DLEITGLQKI FDGSLPMLFN NVKDMPHARA ITNLFGDIRV VEELFGWENS LDRVKKVAR AIDHPLKPVI IGQDEAPVQE EVLTTDLDVN KWLTAIRHTP LETEMTIGSG ISCVVGPYFD GGSHIGYNRM N FRWGNVGT ...String: ERVGEKDLRA ALEWFRSKGY LVETNKEVNP DLEITGLQKI FDGSLPMLFN NVKDMPHARA ITNLFGDIRV VEELFGWENS LDRVKKVAR AIDHPLKPVI IGQDEAPVQE EVLTTDLDVN KWLTAIRHTP LETEMTIGSG ISCVVGPYFD GGSHIGYNRM N FRWGNVGT FQISPGSHMW QVMTEHYKDD EPIPLTMCFG VPPSCTYVAG AGFDYAILPK GCDEIGIAGA IQGSPVRLVK CR TIDAYTL ADAEYVLEGY LHPRDKRYET AESEAADIQG RFHFHPEWAG YMGKAYKAPT FHVTAITMRR RESKPIIFPL GVH TADDAN IDTSVRESAI FALCERLQPG IVQNVHIPYC MTDWGGCIIQ VKKRNQIEEG WQRNFLAAIL ACSQGMRLAI AVSE DVDIY SMDDIMWCLT TRVNPQTDIL NPLPGGRGQT FMPAERMTSG DKQWTASNTQ FEGGMGIDAT VPYGYESDFH RPVYG VDLV KPENFFDAKD IDKMKSRMAG WVLSLARTGR |
-Macromolecule #2: hydroxylated prenyl-FMN
| Macromolecule | Name: hydroxylated prenyl-FMN / type: ligand / ID: 2 / Number of copies: 6 / Formula: BYN |
|---|---|
| Molecular weight | Theoretical: 542.476 Da |
| Chemical component information |  ChemComp-BYN: |
-Macromolecule #3: 2-CN-benzoyl coenzyme A
| Macromolecule | Name: 2-CN-benzoyl coenzyme A / type: ligand / ID: 3 / Number of copies: 6 / Formula: WC8 |
|---|---|
| Molecular weight | Theoretical: 896.65 Da |
-Macromolecule #4: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 4 / Number of copies: 12 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Macromolecule #5: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 5 / Number of copies: 6 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #6: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 6 / Number of copies: 6 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 112 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 135 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 30 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 6250 / Average exposure time: 2.65 sec. / Average electron dose: 50.08 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: experimental model / Details: pdb from the first holo-complex was used |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-8oz5: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)