[English] 日本語
 Yorodumi
Yorodumi- EMDB-17236: CRYO-EM FOCUSED REFINEMENT MAP OF LEISHMANIA MAJOR 80S RIBOSOME :... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CRYO-EM FOCUSED REFINEMENT MAP OF LEISHMANIA MAJOR 80S RIBOSOME : PARENTAL STRAIN | |||||||||
 Map data Map data | Body 1 for focused refinement | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRYO-EM / LEISHMANIA MAJOR / RIBOSOME / TRANSLATION | |||||||||
| Biological species |  Leishmania major (eukaryote) Leishmania major (eukaryote) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Rajan KS / Yonath A | |||||||||
| Funding support |  Israel, European Union, 2 items Israel, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2024 Journal: Cell Rep / Year: 2024Title: Structural and mechanistic insights into the function of Leishmania ribosome lacking a single pseudouridine modification. Authors: K Shanmugha Rajan / Saurav Aryal / Disha-Gajanan Hiregange / Anat Bashan / Hava Madmoni / Mika Olami / Tirza Doniger / Smadar Cohen-Chalamish / Pascal Pescher / Masato Taoka / Yuko Nobe / ...Authors: K Shanmugha Rajan / Saurav Aryal / Disha-Gajanan Hiregange / Anat Bashan / Hava Madmoni / Mika Olami / Tirza Doniger / Smadar Cohen-Chalamish / Pascal Pescher / Masato Taoka / Yuko Nobe / Aliza Fedorenko / Tanaya Bose / Ella Zimermann / Eric Prina / Noa Aharon-Hefetz / Yitzhak Pilpel / Toshiaki Isobe / Ron Unger / Gerald F Späth / Ada Yonath / Shulamit Michaeli /    Abstract: Leishmania is the causative agent of cutaneous and visceral diseases affecting millions of individuals worldwide. Pseudouridine (Ψ), the most abundant modification on rRNA, changes during the ...Leishmania is the causative agent of cutaneous and visceral diseases affecting millions of individuals worldwide. Pseudouridine (Ψ), the most abundant modification on rRNA, changes during the parasite life cycle. Alterations in the level of a specific Ψ in helix 69 (H69) affected ribosome function. To decipher the molecular mechanism of this phenotype, we determine the structure of ribosomes lacking the single Ψ and its parental strain at ∼2.4-3 Å resolution using cryo-EM. Our findings demonstrate the significance of a single Ψ on H69 to its structure and the importance for its interactions with helix 44 and specific tRNAs. Our study suggests that rRNA modification affects translation of mRNAs carrying codon bias due to selective accommodation of tRNAs by the ribosome. Based on the high-resolution structures, we propose a mechanism explaining how the ribosome selects specific tRNAs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17236.map.gz emd_17236.map.gz | 333.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17236-v30.xml emd-17236-v30.xml emd-17236.xml emd-17236.xml | 28.9 KB 28.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17236.png emd_17236.png | 77 KB | ||
| Filedesc metadata |  emd-17236.cif.gz emd-17236.cif.gz | 4.2 KB | ||
| Others |  emd_17236_additional_1.map.gz emd_17236_additional_1.map.gz emd_17236_additional_2.map.gz emd_17236_additional_2.map.gz emd_17236_half_map_1.map.gz emd_17236_half_map_1.map.gz emd_17236_half_map_2.map.gz emd_17236_half_map_2.map.gz | 259.8 MB 253.1 MB 333.7 MB 333.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17236 http://ftp.pdbj.org/pub/emdb/structures/EMD-17236 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17236 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17236 | HTTPS FTP |
-Validation report
| Summary document |  emd_17236_validation.pdf.gz emd_17236_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17236_full_validation.pdf.gz emd_17236_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_17236_validation.xml.gz emd_17236_validation.xml.gz | 17.9 KB | Display | |
| Data in CIF |  emd_17236_validation.cif.gz emd_17236_validation.cif.gz | 21.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17236 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17236 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17236 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17236 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17236.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17236.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Body 1 for focused refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Body 3 for focused refinement
| File | emd_17236_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Body 3 for focused refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
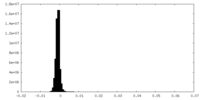
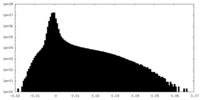
| Density Histograms |
-Additional map: Body 2 for focused refinement
| File | emd_17236_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Body 2 for focused refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
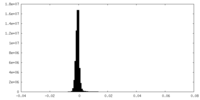
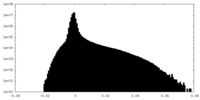
| Density Histograms |
-Half map: Half map2 for Body 1 of focused refinement
| File | emd_17236_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map2 for Body 1 of focused refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map1 for Body 1 of focused refinement
| File | emd_17236_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map1 for Body 1 of focused refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CRYO-EM MAPS OF LEISHMANIA MAJOR 80S RIBOSOME : WILD TYPE REPLICATE 1
| Entire | Name: CRYO-EM MAPS OF LEISHMANIA MAJOR 80S RIBOSOME : WILD TYPE REPLICATE 1 |
|---|---|
| Components |
|
-Supramolecule #1: CRYO-EM MAPS OF LEISHMANIA MAJOR 80S RIBOSOME : WILD TYPE REPLICATE 1
| Supramolecule | Name: CRYO-EM MAPS OF LEISHMANIA MAJOR 80S RIBOSOME : WILD TYPE REPLICATE 1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#85 Details: CRYO-EM MAPS OF LEISHMANIA MAJOR 80S RIBOSOME : WILD TYPE REPLICATE 1 |
|---|---|
| Source (natural) | Organism:  Leishmania major (eukaryote) Leishmania major (eukaryote) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)