[English] 日本語
 Yorodumi
Yorodumi- EMDB-17207: Structure and function of the RAD51B-RAD51C-RAD51D-XRCC2 tumour s... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure and function of the RAD51B-RAD51C-RAD51D-XRCC2 tumour suppressor | ||||||||||||||||||
 Map data Map data | deepEMhancer_map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Complex / ssDNA-binding / ATPase / RAD51 / DNA BINDING PROTEIN | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
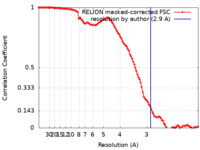
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | ||||||||||||||||||
 Authors Authors | Greenhough LA / Liang CC / West SC | ||||||||||||||||||
| Funding support |  United Kingdom, European Union, 5 items United Kingdom, European Union, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Structure and function of the RAD51B-RAD51C-RAD51D-XRCC2 tumour suppressor. Authors: Luke A Greenhough / Chih-Chao Liang / Ondrej Belan / Simone Kunzelmann / Sarah Maslen / Monica C Rodrigo-Brenni / Roopesh Anand / Mark Skehel / Simon J Boulton / Stephen C West /   Abstract: Homologous recombination is a fundamental process of life. It is required for the protection and restart of broken replication forks, the repair of chromosome breaks and the exchange of genetic ...Homologous recombination is a fundamental process of life. It is required for the protection and restart of broken replication forks, the repair of chromosome breaks and the exchange of genetic material during meiosis. Individuals with mutations in key recombination genes, such as BRCA2 (also known as FANCD1), or the RAD51 paralogues RAD51B, RAD51C (also known as FANCO), RAD51D, XRCC2 (also known as FANCU) and XRCC3, are predisposed to breast, ovarian and prostate cancers and the cancer-prone syndrome Fanconi anaemia. The BRCA2 tumour suppressor protein-the product of BRCA2-is well characterized, but the cellular functions of the RAD51 paralogues remain unclear. Genetic knockouts display growth defects, reduced RAD51 focus formation, spontaneous chromosome abnormalities, sensitivity to PARP inhibitors and replication fork defects, but the precise molecular roles of RAD51 paralogues in fork stability, DNA repair and cancer avoidance remain unknown. Here we used cryo-electron microscopy, AlphaFold2 modelling and structural proteomics to determine the structure of the RAD51B-RAD51C-RAD51D-XRCC2 complex (BCDX2), revealing that RAD51C-RAD51D-XRCC2 mimics three RAD51 protomers aligned within a nucleoprotein filament, whereas RAD51B is highly dynamic. Biochemical and single-molecule analyses showed that BCDX2 stimulates the nucleation and extension of RAD51 filaments-which are essential for recombinational DNA repair-in reactions that depend on the coupled ATPase activities of RAD51B and RAD51C. Our studies demonstrate that BCDX2 orchestrates RAD51 assembly on single stranded DNA for replication fork protection and double strand break repair, in reactions that are critical for tumour avoidance. #1:  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Structure and function of the RAD51B-RAD51C-RAD51D-XRCC2 tumour suppressor Authors: Greenhough LA / Liang CC / Belan O / Kunzelmann S / Maslen S / Rodrigo-Brenni MC / Anand R / Skehel M / Boulton SJ / West SC | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17207.map.gz emd_17207.map.gz | 90.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17207-v30.xml emd-17207-v30.xml emd-17207.xml emd-17207.xml | 22.4 KB 22.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17207_fsc.xml emd_17207_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_17207.png emd_17207.png | 64.1 KB | ||
| Others |  emd_17207_additional_1.map.gz emd_17207_additional_1.map.gz emd_17207_half_map_1.map.gz emd_17207_half_map_1.map.gz emd_17207_half_map_2.map.gz emd_17207_half_map_2.map.gz | 80.9 MB 81 MB 81 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17207 http://ftp.pdbj.org/pub/emdb/structures/EMD-17207 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17207 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17207 | HTTPS FTP |
-Validation report
| Summary document |  emd_17207_validation.pdf.gz emd_17207_validation.pdf.gz | 694.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17207_full_validation.pdf.gz emd_17207_full_validation.pdf.gz | 693.8 KB | Display | |
| Data in XML |  emd_17207_validation.xml.gz emd_17207_validation.xml.gz | 17.6 KB | Display | |
| Data in CIF |  emd_17207_validation.cif.gz emd_17207_validation.cif.gz | 23.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17207 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17207 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17207 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17207 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17207.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17207.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepEMhancer_map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02 Å | ||||||||||||||||||||||||||||||||||||
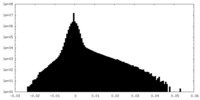
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: refine3D map
| File | emd_17207_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | refine3D_map | ||||||||||||
| Projections & Slices |
| ||||||||||||
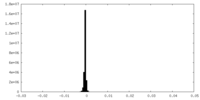
| Density Histograms |
-Half map: half map 1
| File | emd_17207_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
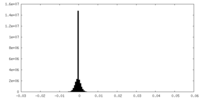
| Density Histograms |
-Half map: half map 2
| File | emd_17207_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
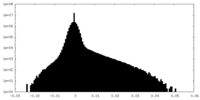
| Density Histograms |
- Sample components
Sample components
-Entire : RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) with 30 nt ssDNA (random sequence)
| Entire | Name: RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) with 30 nt ssDNA (random sequence) |
|---|---|
| Components |
|
-Supramolecule #1: RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) with 30 nt ssDNA (random sequence)
| Supramolecule | Name: RAD51B-RAD51C-RAD51D-XRCC2 (BCDX2) with 30 nt ssDNA (random sequence) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Recombinant BCDX2 complexes purified from Sf9 insect cells, vitrified in the presence of ADP.AlFx |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 25 mM HEPES pH 7.5, 100 mM NaCl, 2.5 mM MgCl2, 0.5 mM ADP.AlFx (0.5 mM ADP, 0.5 mM AlCl3, 10 mM NaF), 0.25 mM TCEP, 0.00075% Tween20, 0.075 mM CHAPSO | ||||||||||||||||||||||||||||||
| Grid | Model: UltrAuFoil R2/2 / Material: GOLD / Mesh: 200 / Support film - Material: GOLD / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: 45 mA | ||||||||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 35305 / Average exposure time: 2.78 sec. / Average electron dose: 53.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.25 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)