+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Partially dissociated CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | CAND1 / substrate receptor exchange factor / cullin-RING ligase / CRL / SCF / ligase / neddylation / DCNL1 co-E3 / ubiquitin signaling | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationSCF complex assembly / positive regulation of protein polyubiquitination / Parkin-FBXW7-Cul1 ubiquitin ligase complex / negative regulation of catalytic activity / F-box domain binding / cellular response to cell-matrix adhesion / Aberrant regulation of mitotic exit in cancer due to RB1 defects / PcG protein complex / cullin-RING-type E3 NEDD8 transferase / NEDD8 transferase activity ...SCF complex assembly / positive regulation of protein polyubiquitination / Parkin-FBXW7-Cul1 ubiquitin ligase complex / negative regulation of catalytic activity / F-box domain binding / cellular response to cell-matrix adhesion / Aberrant regulation of mitotic exit in cancer due to RB1 defects / PcG protein complex / cullin-RING-type E3 NEDD8 transferase / NEDD8 transferase activity / cullin-RING ubiquitin ligase complex / positive regulation of ubiquitin protein ligase activity / cellular response to chemical stress / Cul7-RING ubiquitin ligase complex / ubiquitin-dependent protein catabolic process via the C-end degron rule pathway / Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling / maintenance of protein location in nucleus / positive regulation of protein autoubiquitination / RNA polymerase II transcription initiation surveillance / protein neddylation / cyclin-dependent protein serine/threonine kinase activator activity / NEDD8 ligase activity / VCB complex / negative regulation of response to oxidative stress / Cul5-RING ubiquitin ligase complex / SCF ubiquitin ligase complex / Cul2-RING ubiquitin ligase complex / negative regulation of type I interferon production / ubiquitin-ubiquitin ligase activity / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / Cul4A-RING E3 ubiquitin ligase complex / Cul4-RING E3 ubiquitin ligase complex / Cul3-RING ubiquitin ligase complex / positive regulation of intracellular estrogen receptor signaling pathway / Cul4B-RING E3 ubiquitin ligase complex / ubiquitin ligase complex scaffold activity / cyclin A1-CDK2 complex / cyclin E2-CDK2 complex / cyclin E1-CDK2 complex / negative regulation of mitophagy / cyclin A2-CDK2 complex / positive regulation of DNA-templated DNA replication initiation / Prolactin receptor signaling / G2 Phase / Y chromosome / cyclin-dependent protein kinase activity / Phosphorylation of proteins involved in G1/S transition by active Cyclin E:Cdk2 complexes / positive regulation of heterochromatin formation / p53-Dependent G1 DNA Damage Response / X chromosome / PTK6 Regulates Cell Cycle / regulation of anaphase-promoting complex-dependent catabolic process / Defective binding of RB1 mutants to E2F1,(E2F2, E2F3) / centriole replication / Regulation of APC/C activators between G1/S and early anaphase / telomere maintenance in response to DNA damage / cullin family protein binding / centrosome duplication / G0 and Early G1 / Telomere Extension By Telomerase / positive regulation of RNA polymerase II transcription preinitiation complex assembly / protein K63-linked ubiquitination / Activation of the pre-replicative complex / protein monoubiquitination / ubiquitin ligase complex / cyclin-dependent kinase / cyclin-dependent protein serine/threonine kinase activity / TP53 Regulates Transcription of Genes Involved in G1 Cell Cycle Arrest / ubiquitin-like ligase-substrate adaptor activity / Regulation of MITF-M-dependent genes involved in cell cycle and proliferation / Cajal body / Activation of ATR in response to replication stress / protein K48-linked ubiquitination / Cyclin E associated events during G1/S transition / positive regulation of double-strand break repair via homologous recombination / Cyclin A/B1/B2 associated events during G2/M transition / Nuclear events stimulated by ALK signaling in cancer / Cyclin A:Cdk2-associated events at S phase entry / cyclin-dependent protein kinase holoenzyme complex / condensed chromosome / regulation of G2/M transition of mitotic cell cycle / mitotic G1 DNA damage checkpoint signaling / transcription-coupled nucleotide-excision repair / positive regulation of TORC1 signaling / regulation of cellular response to insulin stimulus / positive regulation of smooth muscle cell proliferation / cellular response to nitric oxide / intrinsic apoptotic signaling pathway / negative regulation of insulin receptor signaling pathway / post-translational protein modification / cyclin binding / regulation of mitotic cell cycle / TBP-class protein binding / Regulation of BACH1 activity / T cell activation / MAP3K8 (TPL2)-dependent MAPK1/3 activation / molecular function activator activity / animal organ morphogenesis / ubiquitin binding / positive regulation of DNA replication Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
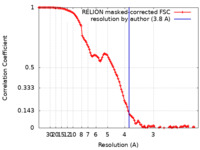
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||||||||
 Authors Authors | Shaaban M / Clapperton JA / Ding S / Maeots ME / Enchev RI / Maslen SL / Skehel JM | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Structural and mechanistic insights into the CAND1-mediated SCF substrate receptor exchange. Authors: Mohammed Shaaban / Julie A Clapperton / Shan Ding / Simone Kunzelmann / Märt-Erik Mäeots / Sarah L Maslen / J Mark Skehel / Radoslav I Enchev /  Abstract: Modular SCF (SKP1-CUL1-Fbox) ubiquitin E3 ligases orchestrate multiple cellular pathways in eukaryotes. Their variable SKP1-Fbox substrate receptor (SR) modules enable regulated substrate recruitment ...Modular SCF (SKP1-CUL1-Fbox) ubiquitin E3 ligases orchestrate multiple cellular pathways in eukaryotes. Their variable SKP1-Fbox substrate receptor (SR) modules enable regulated substrate recruitment and subsequent proteasomal degradation. CAND proteins are essential for the efficient and timely exchange of SRs. To gain structural understanding of the underlying molecular mechanism, we reconstituted a human CAND1-driven exchange reaction of substrate-bound SCF alongside its co-E3 ligase DCNL1 and visualized it by cryo-EM. We describe high-resolution structural intermediates, including a ternary CAND1-SCF complex, as well as conformational and compositional intermediates representing SR- or CAND1-dissociation. We describe in molecular detail how CAND1-induced conformational changes in CUL1/RBX1 provide an optimized DCNL1-binding site and reveal an unexpected dual role for DCNL1 in CAND1-SCF dynamics. Moreover, a partially dissociated CAND1-SCF conformation accommodates cullin neddylation, leading to CAND1 displacement. Our structural findings, together with functional biochemical assays, help formulate a detailed model for CAND-SCF regulation. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17117.map.gz emd_17117.map.gz | 111.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17117-v30.xml emd-17117-v30.xml emd-17117.xml emd-17117.xml | 27.9 KB 27.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17117_fsc.xml emd_17117_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_17117.png emd_17117.png | 124.5 KB | ||
| Filedesc metadata |  emd-17117.cif.gz emd-17117.cif.gz | 8.7 KB | ||
| Others |  emd_17117_half_map_1.map.gz emd_17117_half_map_1.map.gz emd_17117_half_map_2.map.gz emd_17117_half_map_2.map.gz | 98.2 MB 98.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17117 http://ftp.pdbj.org/pub/emdb/structures/EMD-17117 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17117 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17117 | HTTPS FTP |
-Validation report
| Summary document |  emd_17117_validation.pdf.gz emd_17117_validation.pdf.gz | 681.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17117_full_validation.pdf.gz emd_17117_full_validation.pdf.gz | 681.4 KB | Display | |
| Data in XML |  emd_17117_validation.xml.gz emd_17117_validation.xml.gz | 18.7 KB | Display | |
| Data in CIF |  emd_17117_validation.cif.gz emd_17117_validation.cif.gz | 24.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17117 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17117 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17117 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17117 | HTTPS FTP |
-Related structure data
| Related structure data |  8or4MC  8or0C  8or2C  8or3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17117.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17117.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_17117_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
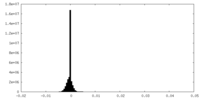
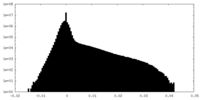
| Density Histograms |
-Half map: #1
| File | emd_17117_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
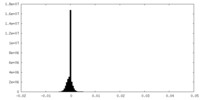
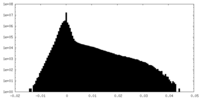
| Density Histograms |
- Sample components
Sample components
-Entire : Partially dissociated CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2
| Entire | Name: Partially dissociated CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 |
|---|---|
| Components |
|
-Supramolecule #1: Partially dissociated CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2
| Supramolecule | Name: Partially dissociated CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Cullin-1
| Macromolecule | Name: Cullin-1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 93.730672 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MWSHPQFEKG SAGSAAGSGA GWSHPQFEKL EVLFQGPMSS TRSQNPHGLK QIGLDQIWDD LRAGIQQVYT RQSMAKSRYM ELYTHVYNY CTSVHQSNQA RGAGVPPSKS KKGQTPGGAQ FVGLELYKRL KEFLKNYLTN LLKDGEDLMD ESVLKFYTQQ W EDYRFSSK ...String: MWSHPQFEKG SAGSAAGSGA GWSHPQFEKL EVLFQGPMSS TRSQNPHGLK QIGLDQIWDD LRAGIQQVYT RQSMAKSRYM ELYTHVYNY CTSVHQSNQA RGAGVPPSKS KKGQTPGGAQ FVGLELYKRL KEFLKNYLTN LLKDGEDLMD ESVLKFYTQQ W EDYRFSSK VLNGICAYLN RHWVRRECDE GRKGIYEIYS LALVTWRDCL FRPLNKQVTN AVLKLIEKER NGETINTRLI SG VVQSYVE LGLNEDDAFA KGPTLTVYKE SFESQFLADT ERFYTRESTE FLQQNPVTEY MKKAEARLLE EQRRVQVYLH EST QDELAR KCEQVLIEKH LEIFHTEFQN LLDADKNEDL GRMYNLVSRI QDGLGELKKL LETHIHNQGL AAIEKCGEAA LNDP KMYVQ TVLDVHKKYN ALVMSAFNND AGFVAALDKA CGRFINNNAV TKMAQSSSKS PELLARYCDS LLKKSSKNPE EAELE DTLN QVMVVFKYIE DKDVFQKFYA KMLAKRLVHQ NSASDDAEAS MISKLKQACG FEYTSKLQRM FQDIGVSKDL NEQFKK HLT NSEPLDLDFS IQVLSSGSWP FQQSCTFALP SELERSYQRF TAFYASRHSG RKLTWLYQLS KGELVTNCFK NRYTLQA ST FQMAILLQYN TEDAYTVQQL TDSTQIKMDI LAQVLQILLK SKLLVLEDEN ANVDEVELKP DTLIKLYLGY KNKKLRVN I NVPMKTEQKQ EQETTHKNIE EDRKLLIQAA IVRIMKMRKV LKHQQLLGEV LTQLSSRFKP RVPVIKKCID ILIEKEYLE RVDGEKDTYS YLA UniProtKB: Cullin-1 |
-Macromolecule #2: E3 ubiquitin-protein ligase RBX1
| Macromolecule | Name: E3 ubiquitin-protein ligase RBX1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.289977 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAAAMDVDTP SGTNSGAGKK RFEVKKWNAV ALWAWDIVVD NCAICRNHIM DLCIECQANQ ASATSEECTV AWGVCNHAFH FHCISRWLK TRQVCPLDNR EWEFQKYGH UniProtKB: E3 ubiquitin-protein ligase RBX1 |
-Macromolecule #3: Cullin-associated NEDD8-dissociated protein 1
| Macromolecule | Name: Cullin-associated NEDD8-dissociated protein 1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 137.358219 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSPEFPGRMA SASYHISNLL EKMTSSDKDF RFMATNDLMT ELQKDSIKLD DDSERKVVKM ILKLLEDKNG EVQNLAVKCL GPLVSKVKE YQVETIVDTL CTNMLSDKEQ LRDISSIGLK TVIGELPPAS SGSALAANVC KKITGRLTSA IAKQEDVSVQ L EALDIMAD ...String: GSPEFPGRMA SASYHISNLL EKMTSSDKDF RFMATNDLMT ELQKDSIKLD DDSERKVVKM ILKLLEDKNG EVQNLAVKCL GPLVSKVKE YQVETIVDTL CTNMLSDKEQ LRDISSIGLK TVIGELPPAS SGSALAANVC KKITGRLTSA IAKQEDVSVQ L EALDIMAD MLSRQGGLLV NFHPSILTCL LPQLTSPRLA VRKRTIIALG HLVMSCGNIV FVDLIEHLLS ELSKNDSMST TR TYIQCIA AISRQAGHRI GEYLEKIIPL VVKFCNVDDD ELREYCIQAF ESFVRRCPKE VYPHVSTIIN ICLKYLTYDP NYN YDDEDE DENAMDADGG DDDDQGSDDE YSDDDDMSWK VRRAAAKCLD AVVSTRHEML PEFYKTVSPA LISRFKEREE NVKA DVFHA YLSLLKQTRP VQSWLCDPDA MEQGETPLTM LQSQVPNIVK ALHKQMKEKS VKTRQCCFNM LTELVNVLPG ALTQH IPVL VPGIIFSLND KSSSSNLKID ALSCLYVILC NHSPQVFHPH VQALVPPVVA CVGDPFYKIT SEALLVTQQL VKVIRP LDQ PSSFDATPYI KDLFTCTIKR LKAADIDQEV KERAISCMGQ IICNLGDNLG SDLPNTLQIF LERLKNEITR LTTVKAL TL IAGSPLKIDL RPVLGEGVPI LASFLRKNQR ALKLGTLSAL DILIKNYSDS LTAAMIDAVL DELPPLISES DMHVSQMA I SFLTTLAKVY PSSLSKISGS ILNELIGLVR SPLLQGGALS AMLDFFQALV VTGTNNLGYM DLLRMLTGPV YSQSTALTH KQSYYSIAKC VAALTRACPK EGPAVVGQFI QDVKNSRSTD SIRLLALLSL GEVGHHIDLS GQLELKSVIL EAFSSPSEEV KSAASYALG SISVGNLPEY LPFVLQEITS QPKRQYLLLH SLKEIISSAS VVGLKPYVEN IWALLLKHCE CAEEGTRNVV A ECLGKLTL IDPETLLPRL KGYLISGSSY ARSSVVTAVK FTISDHPQPI DPLLKNCIGD FLKTLEDPDL NVRRVALVTF NS AAHNKPS LIRDLLDTVL PHLYNETKVR KELIREVEMG PFKHTVDDGL DIRKAAFECM YTLLDSCLDR LDIFEFLNHV EDG LKDHYD IKMLTFLMLV RLSTLCPSAV LQRLDRLVEP LRATCTTKVK ANSVKQEFEK QDELKRSAMR AVAALLTIPE AEKS PLMSE FQSQISSNPE LAAIFESIQK DSSSTNLESM DTS UniProtKB: Cullin-associated NEDD8-dissociated protein 1 |
-Macromolecule #4: S-phase kinase-associated protein 1
| Macromolecule | Name: S-phase kinase-associated protein 1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.679965 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPSIKLQSSD GEIFEVDVEI AKQSVTIKTM LEDLGMDDEG DDDPVPLPNV NAAILKKVIQ WCTHHKDDPP PPEDDENKEK RTDDIPVWD QEFLKVDQGT LFELILAANY LDIKGLLDVT CKTVANMIKG KTPEEIRKTF NIKNDFTEEE EAQVRKENQW C EEK UniProtKB: S-phase kinase-associated protein 1 |
-Macromolecule #5: S-phase kinase-associated protein 2
| Macromolecule | Name: S-phase kinase-associated protein 2 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 48.335312 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSPEFMHRKH LQEIPDLSSN VATSFTWGWD SSKTSELLSG MGVSALEKEE PDSENIPQEL LSNLGHPESP PRKRLKSKGS DKDFVIVRR PKLNRENFPG VSWDSLPDEL LLGIFSCLCL PELLKVSGVC KRWYRLASDE SLWQTLDLTG KNLHPDVTGR L LSQGVIAF ...String: GSPEFMHRKH LQEIPDLSSN VATSFTWGWD SSKTSELLSG MGVSALEKEE PDSENIPQEL LSNLGHPESP PRKRLKSKGS DKDFVIVRR PKLNRENFPG VSWDSLPDEL LLGIFSCLCL PELLKVSGVC KRWYRLASDE SLWQTLDLTG KNLHPDVTGR L LSQGVIAF RCPRSFMDQP LAEHFSPFRV QHMDLSNSVI EVSTLHGILS QCSKLQNLSL EGLRLSDPIV NTLAKNSNLV RL NLSGCSG FSEFALQTLL SSCSRLDELN LSWCFDFTEK HVQVAVAHVS ETITQLNLSG YRKNLQKSDL STLVRRCPNL VHL DLSDSV MLKNDCFQEF FQLNYLQHLS LSRCYDIIPE TLLELGEIPT LKTLQVFGIV PDGTLQLLKE ALPHLQINCS HFTT IARPT IGNKKNQEIW GIKCRLTLQK PSCL UniProtKB: S-phase kinase-associated protein 2 |
-Macromolecule #6: Cyclin-dependent kinases regulatory subunit 1
| Macromolecule | Name: Cyclin-dependent kinases regulatory subunit 1 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.894114 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPLGSQIYYS DKYDDEEFEY RHVMLPKDIA KLVPKTHLMS ESEWRNLGVQ QSQGWVHYMI HEPEPHILLF RRPL UniProtKB: Cyclin-dependent kinases regulatory subunit 1 |
-Macromolecule #7: Cyclin-dependent kinase 2
| Macromolecule | Name: Cyclin-dependent kinase 2 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO / EC number: cyclin-dependent kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 33.976488 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MENFQKVEKI GEGTYGVVYK ARNKLTGEVV ALKKIRLDTE TEGVPSTAIR EISLLKELNH PNIVKLLDVI HTENKLYLVF EFLHQDLKK FMDASALTGI PLPLIKSYLF QLLQGLAFCH SHRVLHRDLK PQNLLINTEG AIKLADFGLA RAFGVPVRTY T HEVVTLWY ...String: MENFQKVEKI GEGTYGVVYK ARNKLTGEVV ALKKIRLDTE TEGVPSTAIR EISLLKELNH PNIVKLLDVI HTENKLYLVF EFLHQDLKK FMDASALTGI PLPLIKSYLF QLLQGLAFCH SHRVLHRDLK PQNLLINTEG AIKLADFGLA RAFGVPVRTY T HEVVTLWY RAPEILLGCK YYSTAVDIWS LGCIFAEMVT RRALFPGDSE IDQLFRIFRT LGTPDEVVWP GVTSMPDYKP SF PKWARQD FSKVVPPLDE DGRSLLSQML HYDPNKRISA KAALAHPFFQ DVTKPVPHLR L UniProtKB: Cyclin-dependent kinase 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | ||||||||||
| Details | CAND1-CUL1-RBX1-SKP1-SKP2-CKS1-CDK2-CyclinE-DCNL1 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 49.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)