+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Double-ring nucleocapsid of the Respiratory Syncytial Virus | |||||||||
 Map data Map data | Double-ring nucleocapsid of the Respiratory Syncytial Virus | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | D10-symmetry / N-RNA / nucleoprotein / RSV / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRespiratory syncytial virus genome transcription / Translation of respiratory syncytial virus mRNAs / symbiont-mediated suppression of host PKR/eIFalpha signaling / helical viral capsid / Respiratory syncytial virus genome replication / RSV-host interactions / Maturation of hRSV A proteins / Assembly and release of respiratory syncytial virus (RSV) virions / Respiratory syncytial virus (RSV) attachment and entry / protein serine/threonine kinase inhibitor activity ...Respiratory syncytial virus genome transcription / Translation of respiratory syncytial virus mRNAs / symbiont-mediated suppression of host PKR/eIFalpha signaling / helical viral capsid / Respiratory syncytial virus genome replication / RSV-host interactions / Maturation of hRSV A proteins / Assembly and release of respiratory syncytial virus (RSV) virions / Respiratory syncytial virus (RSV) attachment and entry / protein serine/threonine kinase inhibitor activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / PKR-mediated signaling / Evasion by RSV of host interferon responses / viral capsid / viral nucleocapsid / symbiont-mediated suppression of host NF-kappaB cascade / host cell cytoplasm / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / ribonucleoprotein complex / RNA binding Similarity search - Function | |||||||||
| Biological species |  Human respiratory syncytial virus A strain Long / Human respiratory syncytial virus A strain Long /  Respiratory syncytial virus / Respiratory syncytial virus /  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Gonnin L / Desfosses A / Gutsche I | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural landscape of the respiratory syncytial virus nucleocapsids. Authors: Lorène Gonnin / Ambroise Desfosses / Maria Bacia-Verloop / Didier Chevret / Marie Galloux / Jean-François Éléouët / Irina Gutsche /  Abstract: Human Respiratory Syncytial Virus (HRSV) is a prevalent cause of severe respiratory infections in children and the elderly. The helical HRSV nucleocapsid is a template for the viral RNA synthesis and ...Human Respiratory Syncytial Virus (HRSV) is a prevalent cause of severe respiratory infections in children and the elderly. The helical HRSV nucleocapsid is a template for the viral RNA synthesis and a scaffold for the virion assembly. This cryo-electron microscopy analysis reveals the non-canonical arrangement of the HRSV nucleocapsid helix, composed of 16 nucleoproteins per asymmetric unit, and the resulting systematic variations in the RNA accessibility. We demonstrate that this unique helical symmetry originates from longitudinal interactions by the C-terminal arm of the HRSV nucleoprotein. We explore the polymorphism of the nucleocapsid-like assemblies, report five structures of the full-length particles and two alternative arrangements formed by a C-terminally truncated nucleoprotein mutant, and demonstrate the functional importance of the identified longitudinal interfaces. We put all these findings in the context of the HRSV RNA synthesis machinery and delineate the structural basis for its further investigation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17031.map.gz emd_17031.map.gz | 7.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17031-v30.xml emd-17031-v30.xml emd-17031.xml emd-17031.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17031.png emd_17031.png | 183.1 KB | ||
| Others |  emd_17031_half_map_1.map.gz emd_17031_half_map_1.map.gz emd_17031_half_map_2.map.gz emd_17031_half_map_2.map.gz | 301.4 MB 301.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17031 http://ftp.pdbj.org/pub/emdb/structures/EMD-17031 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17031 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17031 | HTTPS FTP |
-Validation report
| Summary document |  emd_17031_validation.pdf.gz emd_17031_validation.pdf.gz | 761 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17031_full_validation.pdf.gz emd_17031_full_validation.pdf.gz | 760.6 KB | Display | |
| Data in XML |  emd_17031_validation.xml.gz emd_17031_validation.xml.gz | 16.9 KB | Display | |
| Data in CIF |  emd_17031_validation.cif.gz emd_17031_validation.cif.gz | 19.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17031 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17031 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17031 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17031 | HTTPS FTP |
-Related structure data
| Related structure data |  8oouMC  8op1C  8op2C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17031.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17031.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Double-ring nucleocapsid of the Respiratory Syncytial Virus | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.145 Å | ||||||||||||||||||||||||||||||||||||
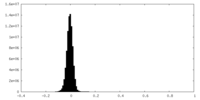
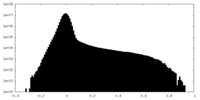
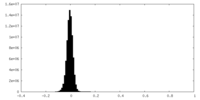
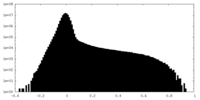
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Double-ring nucleocapsid of the Respiratory Syncytial Virus
| File | emd_17031_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Double-ring nucleocapsid of the Respiratory Syncytial Virus | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Double-ring nucleocapsid of the Respiratory Syncytial Virus
| File | emd_17031_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Double-ring nucleocapsid of the Respiratory Syncytial Virus | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Double ring nucleocapsid of the human Respiratory Syncytial Virus
| Entire | Name: Double ring nucleocapsid of the human Respiratory Syncytial Virus |
|---|---|
| Components |
|
-Supramolecule #1: Double ring nucleocapsid of the human Respiratory Syncytial Virus
| Supramolecule | Name: Double ring nucleocapsid of the human Respiratory Syncytial Virus type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: Double ring nucleocapsid formed by the nucleoprotein N of the human RSV upon overexpression in insect cells and encapsidation of cellular RNA |
|---|---|
| Source (natural) | Organism:  Human respiratory syncytial virus A strain Long Human respiratory syncytial virus A strain Long |
-Macromolecule #1: Nucleoprotein
| Macromolecule | Name: Nucleoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 20 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Respiratory syncytial virus Respiratory syncytial virus |
| Molecular weight | Theoretical: 43.507848 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MALSKVKLND TLNKDQLLSS SKYTIQRSTG DSIDTPNYDV QKHINKLCGM LLITEDANHK FTGLIGMLYA MSRLGREDTI KILRDAGYH VKANGVDVTT HRQDINGKEM KFEVLTLASL TTEIQINIEI ESRKSYKKML KEMGEVAPEY RHDSPDCGMI I LCIAALVI ...String: MALSKVKLND TLNKDQLLSS SKYTIQRSTG DSIDTPNYDV QKHINKLCGM LLITEDANHK FTGLIGMLYA MSRLGREDTI KILRDAGYH VKANGVDVTT HRQDINGKEM KFEVLTLASL TTEIQINIEI ESRKSYKKML KEMGEVAPEY RHDSPDCGMI I LCIAALVI TKLAAGDRSG LTAVIRRANN VLKNEMKRYK GLLPKDIANS FYEVFEKHPH FIDVFVHFGI AQSSTRGGSR VE GIFAGLF MNAYGAGQVM LRWGVLAKSV KNIMLGHASV QAEMEQVVEV YEYAQKLGGE AGFYHILNNP KASLLSLTQF PHF SSVVLG NAAGLGIMGE YRGTPRNQDL YDAAKAYAEQ LKENGVINYS VLDLTAEELE AIKHQLNPKD NDVEL UniProtKB: Nucleoprotein |
-Macromolecule #2: RNA (70-mer)
| Macromolecule | Name: RNA (70-mer) / type: rna / ID: 2 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Molecular weight | Theoretical: 21.317777 KDa |
| Sequence | String: CCCCCCCCCC CCCCCCCCCC CCCCCCCCCC CCCCCCCCCC CCCCCCCCCC CCCCCCCCCC CCCCCCCCCC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.7000000000000001 µm |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D10 (2x10 fold dihedral) / Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 47212 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)