[English] 日本語
 Yorodumi
Yorodumi- EMDB-17001: In-situ structure of the hexameric HEF trimers from influenza C v... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In-situ structure of the hexameric HEF trimers from influenza C viral particles | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Class 1 fusion protein / membrane fusion / viral assembly / hexameric / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationsialate 9-O-acetylesterase activity / sialate 4-O-acetylesterase activity / sialate O-acetylesterase / viral budding from plasma membrane / host cell surface receptor binding / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...sialate 9-O-acetylesterase activity / sialate 4-O-acetylesterase activity / sialate O-acetylesterase / viral budding from plasma membrane / host cell surface receptor binding / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Influenza C virus (C/Johannesburg/1/66) Influenza C virus (C/Johannesburg/1/66) | |||||||||
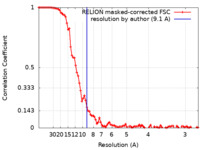
| Method | subtomogram averaging / cryo EM / Resolution: 9.1 Å | |||||||||
 Authors Authors | Liu ZB / Rosenthal PB | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Topological defects in the spherical virus crystal Authors: Liu ZB / Rosenthal PB | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17001.map.gz emd_17001.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17001-v30.xml emd-17001-v30.xml emd-17001.xml emd-17001.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17001_fsc.xml emd_17001_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_17001.png emd_17001.png | 54.3 KB | ||
| Masks |  emd_17001_msk_1.map emd_17001_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17001.cif.gz emd-17001.cif.gz | 5.6 KB | ||
| Others |  emd_17001_half_map_1.map.gz emd_17001_half_map_1.map.gz emd_17001_half_map_2.map.gz emd_17001_half_map_2.map.gz | 48.3 MB 48.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17001 http://ftp.pdbj.org/pub/emdb/structures/EMD-17001 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17001 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17001 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17001.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17001.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38 Å | ||||||||||||||||||||||||||||||||||||
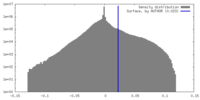
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17001_msk_1.map emd_17001_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
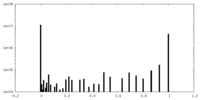
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17001_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
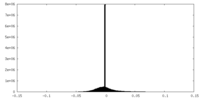
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_17001_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
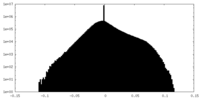
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Influenza C virus (C/Johannesburg/1/66)
| Entire | Name:  Influenza C virus (C/Johannesburg/1/66) Influenza C virus (C/Johannesburg/1/66) |
|---|---|
| Components |
|
-Supramolecule #1: Influenza C virus (C/Johannesburg/1/66)
| Supramolecule | Name: Influenza C virus (C/Johannesburg/1/66) / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Purified viruses / NCBI-ID: 100673 / Sci species name: Influenza C virus (C/Johannesburg/1/66) / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Molecular weight | Theoretical: 1.24 MDa |
-Macromolecule #1: Haemagglutinin-esterase-fusion 1 glycoprotein
| Macromolecule | Name: Haemagglutinin-esterase-fusion 1 glycoprotein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: sialate O-acetylesterase |
|---|---|
| Source (natural) | Organism:  Influenza C virus (C/Johannesburg/1/66) Influenza C virus (C/Johannesburg/1/66) |
| Sequence | String: EKIKICLQKQ VNSSFSLHNG FGGNLYATEE KRMFELVKPK AGASVLNQST WIGFGDSRTD KSNSAFPRSA DVSAKTADKF RFLSGGSLML SMFGPPGKVD YLYQGCGKHK VFYEGVNWSP HAAINCYRKN WTDIKLNFQK NIYELASQSH CMSLVNALDK TIPLQVTAGT ...String: EKIKICLQKQ VNSSFSLHNG FGGNLYATEE KRMFELVKPK AGASVLNQST WIGFGDSRTD KSNSAFPRSA DVSAKTADKF RFLSGGSLML SMFGPPGKVD YLYQGCGKHK VFYEGVNWSP HAAINCYRKN WTDIKLNFQK NIYELASQSH CMSLVNALDK TIPLQVTAGT AGNCNNSFLK NPALYTQEVK PSENKCGKEN LAFFTLPTQF GTYECKLHLV ASCYFIYDSK EVYNKRGCDN YFQVIYDSFG KVVGGLDNRV SPYTGNSGDT PTMQCDMLQL KPGRYSVRSS PRFLLMPERS YCFDMKEKGP VTAVQSIWGK GRESDYAVDQ ACLSTPGCML IQKQKPYIGE ADDHHGDQEM RELLSGLDYE ARCISQSGWV NETSPFTEKY LLPPKFGRCP LAAKEESIPK IPDGLLIPTS GTDTTVTKPK SR UniProtKB: Hemagglutinin-esterase-fusion glycoprotein |
-Macromolecule #2: Haemagglutinin-esterase-fusion 2 glycoprotein
| Macromolecule | Name: Haemagglutinin-esterase-fusion 2 glycoprotein / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO / EC number: sialate O-acetylesterase |
|---|---|
| Source (natural) | Organism:  Influenza C virus (C/Johannesburg/1/66) Influenza C virus (C/Johannesburg/1/66) |
| Sequence | String: IFGIDDLIIG VLFVAIVETG IGGYLLGSRK ESGGGVTKES AEKGFEKIGN DIQILKSSIN IAIEKLNDRI SHDEQAIRDL TLEIENARSE ALLGELGIIR ALLVGNISIG LQESLWELAS EITNRAGDLA VEVSPGCWII DNNICDQSCQ NFIFKFNETA PVPTIPPLDT KIDLQ UniProtKB: Hemagglutinin-esterase-fusion glycoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AMYLAMINE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III | ||||||||||||
| Details | Heterogeneous mix of viral particles, viral like particles and cellular vesicles |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number real images: 1 / Average exposure time: 1.1 sec. / Average electron dose: 2.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)