+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | NAPE-PLD dimer bound to a nanodisc | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NAPE-PLD / Lipid / Membrane / nanodisc / LIPID BINDING PROTEIN | |||||||||
| Biological species |  | |||||||||
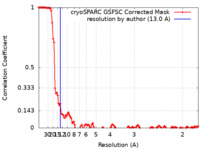
| Method | single particle reconstruction / cryo EM / Resolution: 13.0 Å | |||||||||
 Authors Authors | Chaves-Sanjuan A / Garau G / Bolognesi M | |||||||||
| Funding support |  Italy, 1 items Italy, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Chem Biol / Year: 2025 Journal: Cell Chem Biol / Year: 2025Title: NAPE-PLD is target of thiazide diuretics. Authors: Sara Chiarugi / Francesco Margheriti / Valentina De Lorenzi / Elisa Martino / Eleonora Germana Margheritis / Aldo Moscardini / Roberto Marotta / Antonio Chaves-Sanjuan / Cristina Del Seppia ...Authors: Sara Chiarugi / Francesco Margheriti / Valentina De Lorenzi / Elisa Martino / Eleonora Germana Margheritis / Aldo Moscardini / Roberto Marotta / Antonio Chaves-Sanjuan / Cristina Del Seppia / Giuseppe Federighi / Dominga Lapi / Tiziano Bandiera / Simona Rapposelli / Rossana Scuri / Martino Bolognesi / Gianpiero Garau /  Abstract: Thiazide and thiazide-like diuretics are among the most efficacious and used drugs for the treatment of hypertension, edema, and major cardiovascular outcomes. Despite more then than six decades of ...Thiazide and thiazide-like diuretics are among the most efficacious and used drugs for the treatment of hypertension, edema, and major cardiovascular outcomes. Despite more then than six decades of clinical use, the molecular target and mechanism of action by which these drugs cure hypertension after long-term use have remained mysterious. Here we report the discovery and validation of a previously unknown renal and extrarenal target of these antihypertensives, the membrane-associated phospholipase N-acylphosphatidylethanolamine-specific phospholipase D (NAPE-PLD) of the endocannabinoid system. Structural and functional insights, together with preclinical studies in hypertensive rats, disclose the molecular and physiological basis by which thiazides cause acute diuresis and, at the same time, the distinctive chronic reduction of vascular resistance. Our results shed light on the mechanism of treatment of hypertension and will be useful for developing more efficacious medications for the management of vascular risk factors, as well as associated leukoencephalopathies and myelin disorders. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16931.map.gz emd_16931.map.gz | 85.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16931-v30.xml emd-16931-v30.xml emd-16931.xml emd-16931.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16931_fsc.xml emd_16931_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_16931.png emd_16931.png | 66.6 KB | ||
| Masks |  emd_16931_msk_1.map emd_16931_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16931.cif.gz emd-16931.cif.gz | 4.6 KB | ||
| Others |  emd_16931_half_map_1.map.gz emd_16931_half_map_1.map.gz emd_16931_half_map_2.map.gz emd_16931_half_map_2.map.gz | 165 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16931 http://ftp.pdbj.org/pub/emdb/structures/EMD-16931 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16931 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16931 | HTTPS FTP |
-Validation report
| Summary document |  emd_16931_validation.pdf.gz emd_16931_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16931_full_validation.pdf.gz emd_16931_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_16931_validation.xml.gz emd_16931_validation.xml.gz | 20.5 KB | Display | |
| Data in CIF |  emd_16931_validation.cif.gz emd_16931_validation.cif.gz | 26.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16931 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16931 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16931 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16931 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16931.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16931.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.889 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16931_msk_1.map emd_16931_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16931_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_16931_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NAPE-PLD bound to a nanodisc
| Entire | Name: NAPE-PLD bound to a nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: NAPE-PLD bound to a nanodisc
| Supramolecule | Name: NAPE-PLD bound to a nanodisc / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Details: 30mA | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV / Details: incubated on grid for 60 s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Number grids imaged: 1 / Number real images: 2500 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)