+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human apo pseudouridine synthase 3 (PUS3) | ||||||||||||
 Map data Map data | human full-legnth apo PUS3 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | RNA modification / pseudouridylation / tRNA / homodimer / RNA BINDING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationtRNA pseudouridine38/39 synthase / tRNA pseudouridine(38/39) synthase activity / tRNA pseudouridine synthesis / mRNA pseudouridine synthesis / pseudouridine synthase activity / tRNA modification in the nucleus and cytosol / tRNA modification / RNA binding / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
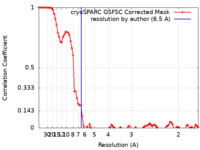
| Method | single particle reconstruction / cryo EM / Resolution: 6.5 Å | ||||||||||||
 Authors Authors | Lin T-Y / Glatt S / Koziej L | ||||||||||||
| Funding support |  Poland, European Union, Poland, European Union,  Switzerland, 3 items Switzerland, 3 items
| ||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: The molecular basis of tRNA selectivity by human pseudouridine synthase 3. Authors: Ting-Yu Lin / Leon Kleemann / Jakub Jeżowski / Dominika Dobosz / Michał Rawski / Paulina Indyka / Grzegorz Ważny / Rahul Mehta / Andrzej Chramiec-Głąbik / Łukasz Koziej / Tristan Ranff ...Authors: Ting-Yu Lin / Leon Kleemann / Jakub Jeżowski / Dominika Dobosz / Michał Rawski / Paulina Indyka / Grzegorz Ważny / Rahul Mehta / Andrzej Chramiec-Głąbik / Łukasz Koziej / Tristan Ranff / Christian Fufezan / Mateusz Wawro / Jakub Kochan / Joanna Bereta / Sebastian A Leidel / Sebastian Glatt /    Abstract: Pseudouridine (Ψ), the isomer of uridine, is ubiquitously found in RNA, including tRNA, rRNA, and mRNA. Human pseudouridine synthase 3 (PUS3) catalyzes pseudouridylation of position 38/39 in tRNAs. ...Pseudouridine (Ψ), the isomer of uridine, is ubiquitously found in RNA, including tRNA, rRNA, and mRNA. Human pseudouridine synthase 3 (PUS3) catalyzes pseudouridylation of position 38/39 in tRNAs. However, the molecular mechanisms by which it recognizes its RNA targets and achieves site specificity remain elusive. Here, we determine single-particle cryo-EM structures of PUS3 in its apo form and bound to three tRNAs, showing how the symmetric PUS3 homodimer recognizes tRNAs and positions the target uridine next to its active site. Structure-guided and patient-derived mutations validate our structural findings in complementary biochemical assays. Furthermore, we deleted PUS1 and PUS3 in HEK293 cells and mapped transcriptome-wide Ψ sites by Pseudo-seq. Although PUS1-dependent sites were detectable in tRNA and mRNA, we found no evidence that human PUS3 modifies mRNAs. Our work provides the molecular basis for PUS3-mediated tRNA modification in humans and explains how its tRNA modification activity is linked to intellectual disabilities. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16917.map.gz emd_16917.map.gz | 30.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16917-v30.xml emd-16917-v30.xml emd-16917.xml emd-16917.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16917_fsc.xml emd_16917_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_16917.png emd_16917.png | 26.8 KB | ||
| Masks |  emd_16917_msk_1.map emd_16917_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16917.cif.gz emd-16917.cif.gz | 5.4 KB | ||
| Others |  emd_16917_half_map_1.map.gz emd_16917_half_map_1.map.gz emd_16917_half_map_2.map.gz emd_16917_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16917 http://ftp.pdbj.org/pub/emdb/structures/EMD-16917 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16917 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16917 | HTTPS FTP |
-Related structure data
| Related structure data |  9f9qMC  8okdC  9enbC  9encC  9eneC  9enfC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16917.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16917.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human full-legnth apo PUS3 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16917_msk_1.map emd_16917_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: human full-legnth apo PUS3 half map A
| File | emd_16917_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human full-legnth apo PUS3 half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: human full-legnth apo PUS3 half map B
| File | emd_16917_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human full-legnth apo PUS3 half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : homodimer of PUS3
| Entire | Name: homodimer of PUS3 |
|---|---|
| Components |
|
-Supramolecule #1: homodimer of PUS3
| Supramolecule | Name: homodimer of PUS3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 550 KDa |
-Macromolecule #1: pseudouridine syntase 3
| Macromolecule | Name: pseudouridine syntase 3 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: tRNA pseudouridine38/39 synthase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: AMADNDTDRN QTEKLLKRVR ELEQEVQRLK KEQAKNKEDS NIRENSAGAG KTKRAFDFSA HGRRHVALRI AYMGWGYQGF ASQENTNNTI EEKLFEALTK TRLVESRQTS NYHRCGRTAK GVSAFGQVIS LDLRSQFPRG RDSEDFNVKE EANAAAEEIR YTHILNRVLP ...String: AMADNDTDRN QTEKLLKRVR ELEQEVQRLK KEQAKNKEDS NIRENSAGAG KTKRAFDFSA HGRRHVALRI AYMGWGYQGF ASQENTNNTI EEKLFEALTK TRLVESRQTS NYHRCGRTAK GVSAFGQVIS LDLRSQFPRG RDSEDFNVKE EANAAAEEIR YTHILNRVLP PDIRILAWAP VEPSFSARFS CLERTYRYFF PRADLDIVTM DYAAQKYVGT HDFRNLCKMD VANGVINFQR TILSAQVQLV GQSPGEGRWQ EPFQLCQFEV TGQAFLYHQV RCMMAILFLI GQGMEKPEII DELLNIEKNP QKPQYSMAVE FPLVLYDCKF ENVKWIYDQE AQEFNITHLQ QLWANHAVKT HMLYSMLQGL DTVPVPCGIG PKMDGMTEWG NVKPSVIKQT SAFVEGVKMR TYKPLMDRPK CQGLESRIQH FVRRGRIEHP HLFHEEETKA KRDCNDTLEE ENTNLETPTK RVCVDTEIKS II UniProtKB: tRNA pseudouridine(38/39) synthase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: C-flat-2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: 8 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 7353 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.8000000000000003 µm / Nominal defocus min: 3.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)