登録情報 データベース : EMDB / ID : EMD-16842タイトル SIRT6 bound nucleosome Sharpened map 複合体 : Human Sirtuin 6 in complex with the nucleosome複合体 : Human NAD-dependent protein deacylase sirtuin-6タンパク質・ペプチド : Human NAD-dependent protein deacylase sirtuin-6複合体 : Histonesタンパク質・ペプチド : Histone H2Aタンパク質・ペプチド : Histone H2Bタンパク質・ペプチド : Histone H3タンパク質・ペプチド : Histone H4複合体 : DNADNA : DNA (145-MER)DNA : DNA (145-MER) / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
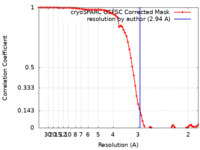
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Homo sapiens (ヒト) / Xenopus laevis (アフリカツメガエル) / synthetic construct (人工物) 手法 / / 解像度 : 2.94 Å Smirnova E / Bignon E / Schultz P / Papai G / Ben-Shem A 資金援助 Organization Grant number 国 Other private
ジャーナル : Elife / 年 : 2024タイトル : Binding to nucleosome poises human SIRT6 for histone H3 deacetylation.
著者 :
Ekaterina Smirnova / Emmanuelle Bignon / Patrick Schultz / Gabor Papai / Adam Ben Shem / 要旨 :
Sirtuin 6 (SIRT6) is an NAD-dependent histone H3 deacetylase that is prominently found associated with chromatin, attenuates transcriptionally active promoters and regulates DNA repair, metabolic ... Sirtuin 6 (SIRT6) is an NAD-dependent histone H3 deacetylase that is prominently found associated with chromatin, attenuates transcriptionally active promoters and regulates DNA repair, metabolic homeostasis and lifespan. Unlike other sirtuins, it has low affinity to free histone tails but demonstrates strong binding to nucleosomes. It is poorly understood how SIRT6 docking on nucleosomes stimulates its histone deacetylation activity. Here, we present the structure of human SIRT6 bound to a nucleosome determined by cryogenic electron microscopy. The zinc finger domain of SIRT6 associates tightly with the acidic patch of the nucleosome through multiple arginine anchors. The Rossmann fold domain binds to the terminus of the looser DNA half of the nucleosome, detaching two turns of the DNA from the histone octamer and placing the NAD binding pocket close to the DNA exit site. This domain shows flexibility with respect to the fixed zinc finger and moves with, but also relative to, the unwrapped DNA terminus. We apply molecular dynamics simulations of the histone tails in the nucleosome to show that in this mode of interaction, the active site of SIRT6 is perfectly poised to catalyze deacetylation of the H3 histone tail and that the partial unwrapping of the DNA allows even lysines close to the H3 core to reach the enzyme. 履歴 登録 2023年3月13日 - ヘッダ(付随情報) 公開 2023年8月9日 - マップ公開 2023年8月9日 - 更新 2024年3月13日 - 現状 2024年3月13日 処理サイト : PDBe / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報
 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト) /
Homo sapiens (ヒト) /  データ登録者
データ登録者 フランス, 1件
フランス, 1件  引用
引用 ジャーナル: Elife / 年: 2024
ジャーナル: Elife / 年: 2024
 ジャーナル: Elife / 年: 2023
ジャーナル: Elife / 年: 2023 構造の表示
構造の表示 ダウンロードとリンク
ダウンロードとリンク emd_16842.map.gz
emd_16842.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-16842-v30.xml
emd-16842-v30.xml emd-16842.xml
emd-16842.xml EMDBヘッダ
EMDBヘッダ emd_16842_fsc.xml
emd_16842_fsc.xml FSCデータファイル
FSCデータファイル emd_16842.png
emd_16842.png emd_16842_msk_1.map
emd_16842_msk_1.map マスクマップ
マスクマップ emd-16842.cif.gz
emd-16842.cif.gz emd_16842_half_map_1.map.gz
emd_16842_half_map_1.map.gz emd_16842_half_map_2.map.gz
emd_16842_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-16842
http://ftp.pdbj.org/pub/emdb/structures/EMD-16842 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16842
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16842 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_16842.map.gz / 形式: CCP4 / 大きさ: 42.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_16842.map.gz / 形式: CCP4 / 大きさ: 42.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) emd_16842_msk_1.map
emd_16842_msk_1.map 試料の構成要素
試料の構成要素 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 ムービー
ムービー コントローラー
コントローラー














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)