[English] 日本語
 Yorodumi
Yorodumi- EMDB-16746: High resolution structure of the coagulation Factor XIII A2B2 het... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | High resolution structure of the coagulation Factor XIII A2B2 heterotetramer complex. | |||||||||
 Map data Map data | High-resolution reconstruction of Factor XIII A2B2 with C2 symmetry. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Factor XIII / BLOOD CLOTTING | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein-glutamine gamma-glutamyltransferase / protein-glutamine gamma-glutamyltransferase activity / transferase complex / peptide cross-linking / blood coagulation, fibrin clot formation / Common Pathway of Fibrin Clot Formation / platelet alpha granule lumen / : / blood coagulation / Platelet degranulation ...protein-glutamine gamma-glutamyltransferase / protein-glutamine gamma-glutamyltransferase activity / transferase complex / peptide cross-linking / blood coagulation, fibrin clot formation / Common Pathway of Fibrin Clot Formation / platelet alpha granule lumen / : / blood coagulation / Platelet degranulation / Interleukin-4 and Interleukin-13 signaling / blood microparticle / extracellular space / extracellular region / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
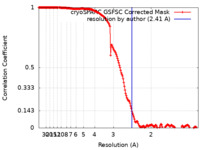
| Method | single particle reconstruction / cryo EM / Resolution: 2.41 Å | |||||||||
 Authors Authors | Singh S / Urgular D / Hagelueken G / Geyer M / Biswas A | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Blood / Year: 2025 Journal: Blood / Year: 2025Title: Cryo-EM structure of the human native plasma coagulation factor XIII complex. Authors: Sneha Singh / Gregor Hagelueken / Deniz Ugurlar / Samhitha Urs Ramaraje Urs / Amit Sharma / Manoranjan Mahapatra / Friedel Drepper / Diana Imhof / Pitter F Huesgen / Johannes Oldenburg / ...Authors: Sneha Singh / Gregor Hagelueken / Deniz Ugurlar / Samhitha Urs Ramaraje Urs / Amit Sharma / Manoranjan Mahapatra / Friedel Drepper / Diana Imhof / Pitter F Huesgen / Johannes Oldenburg / Matthias Geyer / Arijit Biswas /    Abstract: The structure of human coagulation factor XIII (FXIII), a heterotetrameric plasma protransglutaminase that covalently cross-links preformed fibrin polymers, remains elusive until today. The ...The structure of human coagulation factor XIII (FXIII), a heterotetrameric plasma protransglutaminase that covalently cross-links preformed fibrin polymers, remains elusive until today. The heterotetrameric complex is composed of 2 catalytic FXIII-A and 2 protective FXIII-B subunits. Structural etiology underlying FXIII deficiency has so far been derived from crystallographic structures, all of which are currently available for the FXIII-A2 homodimer only. Here, we present the cryogenic electron microscopy (cryo-EM) structure of a native, human plasma-derived FXIII-A2B2 complex at 2.4 Å resolution. The structure provides detailed information on FXIII subunit interacting interfaces as the 2 subunits interact strongly in plasma. The native FXIII-A2B2 complex reveals a pseudosymmetric heterotetramer of 2 FXIII-B monomers intercalating with a symmetric FXIII-A2 dimer forming a "crown"-like assembly. The symmetry axes of the A2 and B2 homodimers are twisted relative to each other such that Sushi domain 1 interacts with the catalytic core of the A subunit, and Sushi domain 2 with the symmetry related A' subunit, and vice versa. We also report 4 novel mutations in the F13A1 gene encoding the FXIII-A subunit from a cohort of patients with severe FXIII deficiency. Our structure reveals the etiological basis of homozygous and heterozygous pathogenic mutations and explains the conditional dominant negative effects of heterozygous mutations. This atomistic description of complex interfaces is consistent with previous biochemical data and shows a congruence between the structural biochemistry of the FXIII complex and the clinical features of FXIII deficiency. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16746.map.gz emd_16746.map.gz | 483.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16746-v30.xml emd-16746-v30.xml emd-16746.xml emd-16746.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16746_fsc.xml emd_16746_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_16746.png emd_16746.png | 77.9 KB | ||
| Filedesc metadata |  emd-16746.cif.gz emd-16746.cif.gz | 6.3 KB | ||
| Others |  emd_16746_half_map_1.map.gz emd_16746_half_map_1.map.gz emd_16746_half_map_2.map.gz emd_16746_half_map_2.map.gz | 474.3 MB 474.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16746 http://ftp.pdbj.org/pub/emdb/structures/EMD-16746 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16746 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16746 | HTTPS FTP |
-Related structure data
| Related structure data |  8cmuMC  8cmtC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16746.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16746.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | High-resolution reconstruction of Factor XIII A2B2 with C2 symmetry. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.697 Å | ||||||||||||||||||||||||||||||||||||
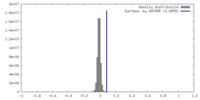
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map A
| File | emd_16746_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
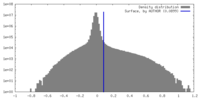
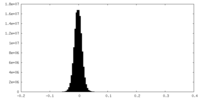
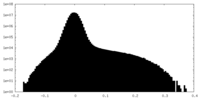
| Density Histograms |
-Half map: Half map B
| File | emd_16746_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Factor XIII A2B2 heterotetramer
| Entire | Name: Factor XIII A2B2 heterotetramer |
|---|---|
| Components |
|
-Supramolecule #1: Factor XIII A2B2 heterotetramer
| Supramolecule | Name: Factor XIII A2B2 heterotetramer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Coagulation factor XIII A chain
| Macromolecule | Name: Coagulation factor XIII A chain / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: protein-glutamine gamma-glutamyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 83.365109 KDa |
| Sequence | String: MSETSRTAFG GRRAVPPNNS NAAEDDLPTV ELQGVVPRGV NLQEFLNVTS VHLFKERWDT NKVDHHTDKY ENNKLIVRRG QSFYVQIDF SRPYDPRRDL FRVEYVIGRY PQENKGTYIP VPIVSELQSG KWGAKIVMRE DRSVRLSIQS SPKCIVGKFR M YVAVWTPY ...String: MSETSRTAFG GRRAVPPNNS NAAEDDLPTV ELQGVVPRGV NLQEFLNVTS VHLFKERWDT NKVDHHTDKY ENNKLIVRRG QSFYVQIDF SRPYDPRRDL FRVEYVIGRY PQENKGTYIP VPIVSELQSG KWGAKIVMRE DRSVRLSIQS SPKCIVGKFR M YVAVWTPY GVLRTSRNPE TDTYILFNPW CEDDAVYLDN EKEREEYVLN DIGVIFYGEV NDIKTRSWSY GQFEDGILDT CL YVMDRAQ MDLSGRGNPI KVSRVGSAMV NAKDDEGVLV GSWDNIYAYG VPPSAWTGSV DILLEYRSSE NPVRYGQCWV FAG VFNTFL RCLGIPARIV TNYFSAHDND ANLQMDIFLE EDGNVNSKLT KDSVWNYHCW NEAWMTRPDL PVGFGGWQAV DSTP QENSD GMYRCGPASV QAIKHGHVCF QFDAPFVFAE VNSDLIYITA KKDGTHVVEN VDATHIGKLI VTKQIGGDGM MDITD TYKF QEGQEEERLA LETALMYGAK KPLNTEGVMK SRSNVDMDFE VENAVLGKDF KLSITFRNNS HNRYTITAYL SANITF YTG VPKAEFKKET FDVTLEPLSF KKEAVLIQAG EYMGQLLEQA SLHFFVTARI NETRDVLAKQ KSTVLTIPEI IIKVRGT QV VGSDMTVTVE FTNPLKETLR NVWVHLDGPG VTRPMKKMFR EIRPNSTVQW EEVCRPWVSG HRKLIASMSS DSLRHVYG E LDVQIQRRPS M UniProtKB: Coagulation factor XIII A chain |
-Macromolecule #2: Coagulation factor XIII B chain
| Macromolecule | Name: Coagulation factor XIII B chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 75.600594 KDa |
| Sequence | String: MRLKNLTFII ILIISGELYA EEKPCGFPHV ENGRIAQYYY TFKSFYFPMS IDKKLSFFCL AGYTTESGRQ EEQTTCTTEG WSPEPRCFK KCTKPDLSNG YISDVKLLYK IQENMRYGCA SGYKTTGGKD EEVVQCLSDG WSSQPTCRKE HETCLAPELY N GNYSTTQK ...String: MRLKNLTFII ILIISGELYA EEKPCGFPHV ENGRIAQYYY TFKSFYFPMS IDKKLSFFCL AGYTTESGRQ EEQTTCTTEG WSPEPRCFK KCTKPDLSNG YISDVKLLYK IQENMRYGCA SGYKTTGGKD EEVVQCLSDG WSSQPTCRKE HETCLAPELY N GNYSTTQK TFKVKDKVQY ECATGYYTAG GKKTEEVECL TYGWSLTPKC TKLKCSSLRL IENGYFHPVK QTYEEGDVVQ FF CHENYYL SGSDLIQCYN FGWYPESPVC EGRRNRCPPP PLPINSKIQT HSTTYRHGEI VHIECELNFE IHGSAEIRCE DGK WTEPPK CIEGQEKVAC EEPPFIENGA ANLHSKIYYN GDKVTYACKS GYLLHGSNEI TCNRGKWTLP PECVENNENC KHPP VVMNG AVADGILASY ATGSSVEYRC NEYYLLRGSK ISRCEQGKWS SPPVCLEPCT VNVDYMNRNN IEMKWKYEGK VLHGD LIDF VCKQGYDLSP LTPLSELSVQ CNRGEVKYPL CTRKESKGMC TSPPLIKHGV IISSTVDTYE NGSSVEYRCF DHHFLE GSR EAYCLDGMWT TPPLCLEPCT LSFTEMEKNN LLLKWDFDNR PHILHGEYIE FICRGDTYPA ELYITGSILR MQCDRGQ LK YPRCIPRQST LSYQEPLRT UniProtKB: Coagulation factor XIII B chain |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 748 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)