[English] 日本語
 Yorodumi
Yorodumi- EMDB-16731: Alpha-ketoglutarate dehydrogenase complex core (E2, dihydrolipoyl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Alpha-ketoglutarate dehydrogenase complex core (E2, dihydrolipoyl transsuccinylase), Branched-chain alpha-ketoacid dehydrogenase complex core (E2, dihydrolipoyl transacylase), indistinguishable | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transferase / cube / mitochondrion | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 32.0 Å | |||||||||
 Authors Authors | Plokhikh KS / Chesnokov YM | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: FEBS J / Year: 2024 Journal: FEBS J / Year: 2024Title: Association of 2-oxoacid dehydrogenase complexes with respirasomes in mitochondria. Authors: Konstantin S Plokhikh / Semen V Nesterov / Yuriy M Chesnokov / Anton G Rogov / Roman A Kamyshinsky / Aleksandr L Vasiliev / Lev S Yaguzhinsky / Raif G Vasilov /  Abstract: In the present study, cryo-electron tomography was used to investigate the localization of 2-oxoacid dehydrogenase complexes (OADCs) in cardiac mitochondria and mitochondrial inner membrane samples. ...In the present study, cryo-electron tomography was used to investigate the localization of 2-oxoacid dehydrogenase complexes (OADCs) in cardiac mitochondria and mitochondrial inner membrane samples. Two classes of ordered OADC inner cores with different symmetries were distinguished and their quaternary structures modeled. One class corresponds to pyruvate dehydrogenase complexes and the other to dehydrogenase complexes of α-ketoglutarate and branched-chain α-ketoacids. OADCs were shown to be localized in close proximity to membrane-embedded respirasomes, as observed both in densely packed lamellar cristae of cardiac mitochondria and in ruptured mitochondrial samples where the dense packing is absent. This suggests the specificity of the OADC-respirasome interaction, which allows localized NADH/NAD exchange between OADCs and complex I of the respiratory chain. The importance of this local coupling is based on OADCs being the link between respiration, glycolysis and amino acid metabolism. The coupling of these basic metabolic processes can vary in different tissues and conditions and may be involved in the development of various pathologies. The present study shows that this important and previously missing parameter of mitochondrial complex coupling can be successfully assessed using cryo-electron tomography. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16731.map.gz emd_16731.map.gz | 367 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16731-v30.xml emd-16731-v30.xml emd-16731.xml emd-16731.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16731_fsc.xml emd_16731_fsc.xml | 6 KB | Display |  FSC data file FSC data file |
| Images |  emd_16731.png emd_16731.png | 166.4 KB | ||
| Masks |  emd_16731_msk_1.map emd_16731_msk_1.map | 8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16731.cif.gz emd-16731.cif.gz | 4.2 KB | ||
| Others |  emd_16731_half_map_1.map.gz emd_16731_half_map_1.map.gz emd_16731_half_map_2.map.gz emd_16731_half_map_2.map.gz | 4.6 MB 4.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16731 http://ftp.pdbj.org/pub/emdb/structures/EMD-16731 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16731 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16731 | HTTPS FTP |
-Validation report
| Summary document |  emd_16731_validation.pdf.gz emd_16731_validation.pdf.gz | 502.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16731_full_validation.pdf.gz emd_16731_full_validation.pdf.gz | 501.8 KB | Display | |
| Data in XML |  emd_16731_validation.xml.gz emd_16731_validation.xml.gz | 10.6 KB | Display | |
| Data in CIF |  emd_16731_validation.cif.gz emd_16731_validation.cif.gz | 14.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16731 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16731 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16731 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16731 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16731.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16731.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.7 Å | ||||||||||||||||||||||||||||||||||||
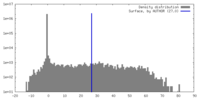
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16731_msk_1.map emd_16731_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
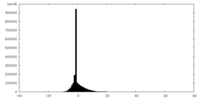
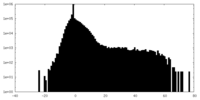
| Density Histograms |
-Half map: #1
| File | emd_16731_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
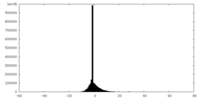
| Density Histograms |
-Half map: #2
| File | emd_16731_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
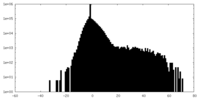
| Density Histograms |
- Sample components
Sample components
-Entire : ruptured mitochondrial membrane samples
| Entire | Name: ruptured mitochondrial membrane samples |
|---|---|
| Components |
|
-Supramolecule #1: ruptured mitochondrial membrane samples
| Supramolecule | Name: ruptured mitochondrial membrane samples / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Component - Concentration: 10.0 mM / Component - Name: Hepes/KOH |
|---|---|
| Grid | Model: EMS Lacey Carbon / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 1.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 8.0 µm / Nominal defocus min: 6.0 µm / Nominal magnification: 18000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)