+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1670 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a rabbit ribosome with P site tRNA | |||||||||
 Map data Map data | This is a map of a rabbit reticulocyte lysate ribosome stalled on a mRNA by a stem-loop with a P-site tRNA. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome / mRNA / tRNA / stem-loop | |||||||||
| Biological species |   Infectious bronchitis virus Infectious bronchitis virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 13.6 Å | |||||||||
 Authors Authors | Flanagan IV JF / Namy O / Brierley I / Gilbert RJC | |||||||||
 Citation Citation |  Journal: Structure / Year: 2010 Journal: Structure / Year: 2010Title: Direct observation of distinct A/P hybrid-state tRNAs in translocating ribosomes. Authors: John F Flanagan / Olivier Namy / Ian Brierley / Robert J C Gilbert /   Abstract: Transfer RNAs (tRNAs) link the genetic code in the form of messenger RNA (mRNA) to protein sequence. Translocation of tRNAs through the ribosome from aminoacyl (A) site to peptidyl (P) site and from ...Transfer RNAs (tRNAs) link the genetic code in the form of messenger RNA (mRNA) to protein sequence. Translocation of tRNAs through the ribosome from aminoacyl (A) site to peptidyl (P) site and from P site to exit site is catalyzed in eukaryotes by the translocase elongation factor 2 (EF-2) and in prokaryotes by its homolog EF-G. During tRNA movement one or more "hybrid" states (A/P) is occupied, but molecular details of them and of the translocation process are limited. Here we show by cryo-electron microscopy that a population of mammalian ribosomes stalled at an mRNA pseudoknot structure contains structurally distorted tRNAs in two different A/P hybrid states. In one (A/P'), the tRNA is in contact with the translocase EF-2, which induces it. In the other (A/P''), the translocase is absent. The existence of these alternative A/P intermediate states has relevance to our understanding of the mechanics and kinetics of translocation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1670.map.gz emd_1670.map.gz | 11.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1670-v30.xml emd-1670-v30.xml emd-1670.xml emd-1670.xml | 11.3 KB 11.3 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-1670.gif EMD-1670.gif | 83.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1670 http://ftp.pdbj.org/pub/emdb/structures/EMD-1670 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1670 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1670 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1670.map.gz / Format: CCP4 / Size: 96.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1670.map.gz / Format: CCP4 / Size: 96.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a map of a rabbit reticulocyte lysate ribosome stalled on a mRNA by a stem-loop with a P-site tRNA. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
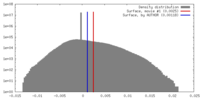
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Rabbit reticulocyte ribosome stalled on a stem-loop-containing mR...
| Entire | Name: Rabbit reticulocyte ribosome stalled on a stem-loop-containing mRNA with a P-site tRNA |
|---|---|
| Components |
|
-Supramolecule #1000: Rabbit reticulocyte ribosome stalled on a stem-loop-containing mR...
| Supramolecule | Name: Rabbit reticulocyte ribosome stalled on a stem-loop-containing mRNA with a P-site tRNA type: sample / ID: 1000 / Oligomeric state: Ribosome, mRNA and tRNA / Number unique components: 3 |
|---|
-Supramolecule #1: 80S ribosome
| Supramolecule | Name: 80S ribosome / type: organelle_or_cellular_component / ID: 1 / Name.synonym: Ribosome Details: Sample was purified by affinity chromatography using a tag in the mRNA. Number of copies: 1 / Oligomeric state: 40S and 60S / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: tRNA
| Macromolecule | Name: tRNA / type: rna / ID: 1 / Name.synonym: tRNA Details: Sample was purified by affinity chromatography using a tag in the mRNA. Classification: TRANSFER / Structure: OTHER / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #2: Infectious bronchitis virus mRNA
| Macromolecule | Name: Infectious bronchitis virus mRNA / type: rna / ID: 2 / Name.synonym: mRNA / Details: Sample was purified by affinity chromatography. / Classification: OTHER / Structure: SINGLE STRANDED / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  Infectious bronchitis virus Infectious bronchitis virus |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 Details: 25mM Hepes-KOH pH 7.8, 5mM MgOAc, 1mM cycloheximide, 150mM KOAc |
|---|---|
| Grid | Details: 300 mesh copper grid with lacey carbon film |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 100 K / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: Homemade plunger / Method: Blot with Whatman number 1 paper |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Temperature | Average: 115 K |
| Alignment procedure | Legacy - Astigmatism: At 200,000 magnification |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 65 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 3.7 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 38000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Per micrograph |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 13.6 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Spider, WellMAP Details: Maps were calculated using 24738 out of 38720 particles selected by the P-site tRNA presence. Presence of the tRNA was proved using multivariate statistical analysis focused classification ...Details: Maps were calculated using 24738 out of 38720 particles selected by the P-site tRNA presence. Presence of the tRNA was proved using multivariate statistical analysis focused classification of particles without previous alignment to a tRNA-containing ribosome. Number images used: 24738 |
| Final angle assignment | Details: Spider convention |
-Atomic model buiding 1
| Initial model | PDB ID:  2zkq |
|---|---|
| Software | Name: URO |
| Details | Protocol: Rigid body in segments. Subregions were defined as described in the paper accompanying this deposition. |
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
-Atomic model buiding 2
| Initial model | PDB ID:  2zkr |
|---|---|
| Software | Name: URO |
| Details | Protocol: Rigid body in segments. Subregions were defined as described in the paper accompanying this deposition. |
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT / Target criteria: correlation coefficient |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)