[English] 日本語
 Yorodumi
Yorodumi- EMDB-16695: Tomogram of an induced protrusion of a Drosophila S2 alpha-tubuli... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tomogram of an induced protrusion of a Drosophila S2 alpha-tubulin acetyltransferase knock-out (dTAT KO) cell with a filament inside the microtubule lumen | |||||||||
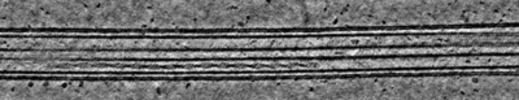
 Map data Map data | Deconvolved tomogram (binned by four) of an induced protrusion from Drosophila S2 alpha-tubulin acetyltransferase knock-out (dTAT KO) cells. From Dataset 8. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Ventura Santos C / Carter AP | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: CryoET shows cofilactin filaments inside the microtubule lumen. Authors: Camilla Ventura Santos / Stephen L Rogers / Andrew P Carter /  Abstract: Cytoplasmic microtubules are tubular polymers that can harbor small proteins or filaments inside their lumen. The identity of these objects and what causes their accumulation has not been ...Cytoplasmic microtubules are tubular polymers that can harbor small proteins or filaments inside their lumen. The identity of these objects and what causes their accumulation has not been conclusively established. Here, we used cryogenic electron tomography (cryoET) of S2 cell protrusions and found filaments inside the microtubule lumen, which resemble those reported recently in human HAP1 cells. The frequency of these filaments increased upon inhibition of the sarco/endoplasmic reticulum Ca ATPase (SERCA) with the small-molecule drug thapsigargin. Subtomogram averaging showed that the luminal filaments adopt a helical structure reminiscent of cofilin-bound actin (cofilactin). Consistent with this, cofilin was activated in cells under the same conditions that increased luminal filament occurrence. Furthermore, RNAi knock-down of cofilin reduced the frequency of luminal filaments with cofilactin morphology. These results suggest that cofilin activation stimulates its accumulation on actin filaments inside the microtubule lumen. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16695.map.gz emd_16695.map.gz | 1.2 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16695-v30.xml emd-16695-v30.xml emd-16695.xml emd-16695.xml | 9.3 KB 9.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_16695.png emd_16695.png | 43.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16695 http://ftp.pdbj.org/pub/emdb/structures/EMD-16695 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16695 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16695 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16695.map.gz / Format: CCP4 / Size: 1.2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16695.map.gz / Format: CCP4 / Size: 1.2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Deconvolved tomogram (binned by four) of an induced protrusion from Drosophila S2 alpha-tubulin acetyltransferase knock-out (dTAT KO) cells. From Dataset 8. | ||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 11.808 Å | ||||||||||||||||||||||||||||||||
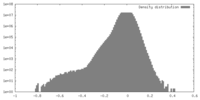
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Tomogram of an induced protrusion of a Drosophila S2 alpha-tublin...
| Entire | Name: Tomogram of an induced protrusion of a Drosophila S2 alpha-tublin acetyltransferase knock-out cell. |
|---|---|
| Components |
|
-Supramolecule #1: Tomogram of an induced protrusion of a Drosophila S2 alpha-tublin...
| Supramolecule | Name: Tomogram of an induced protrusion of a Drosophila S2 alpha-tublin acetyltransferase knock-out cell. type: cell / ID: 1 / Parent: 0 Details: Protrusion formation was induced by treatment with 5 uM Cytochalasin D for 4h. Example tomogram from Dataset 8. |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 298.15 K / Instrument: FEI VITROBOT MARK III |
| Details | Cell were treated with 5 uM Cytochalasin D for 4 h before vitrification. |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: BBI Solutions / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 3.0 e/Å2 Details: Data was collected on Gatan K2 summit (2.952 A/pixel) with a total dose of 122.3 e/A2. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 6.0 µm / Nominal defocus min: 2.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number images used: 41 |
|---|
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)