+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM Structure of Craf:14-3-3:Mek1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Kinase / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationdeath-inducing signaling complex assembly / epithelial cell proliferation involved in lung morphogenesis / positive regulation of endodermal cell differentiation / negative regulation of homotypic cell-cell adhesion / negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway / regulation of vascular associated smooth muscle contraction / intermediate filament cytoskeleton organization / regulation of axon regeneration / mitogen-activated protein kinase kinase / positive regulation of muscle contraction ...death-inducing signaling complex assembly / epithelial cell proliferation involved in lung morphogenesis / positive regulation of endodermal cell differentiation / negative regulation of homotypic cell-cell adhesion / negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway / regulation of vascular associated smooth muscle contraction / intermediate filament cytoskeleton organization / regulation of axon regeneration / mitogen-activated protein kinase kinase / positive regulation of muscle contraction / Golgi inheritance / placenta blood vessel development / MAP-kinase scaffold activity / cerebellar cortex formation / labyrinthine layer development / regulation of Rho protein signal transduction / melanosome transport / type B pancreatic cell proliferation / Signaling by MAP2K mutants / SHOC2 M1731 mutant abolishes MRAS complex function / Gain-of-function MRAS complexes activate RAF signaling / Rap1 signalling / vesicle transport along microtubule / insulin secretion involved in cellular response to glucose stimulus / positive regulation of Ras protein signal transduction / regulation of Golgi inheritance / central nervous system neuron differentiation / mitogen-activated protein kinase kinase kinase binding / positive regulation of axonogenesis / trachea formation / triglyceride homeostasis / Negative feedback regulation of MAPK pathway / regulation of early endosome to late endosome transport / IFNG signaling activates MAPKs / regulation of stress-activated MAPK cascade / GP1b-IX-V activation signalling / Frs2-mediated activation / MAPK3 (ERK1) activation / ERBB2-ERBB3 signaling pathway / neurotrophin TRK receptor signaling pathway / face development / regulation of neurotransmitter receptor localization to postsynaptic specialization membrane / endodermal cell differentiation / pseudopodium / MAP kinase kinase activity / Bergmann glial cell differentiation / positive regulation of ATP biosynthetic process / regulation of cell differentiation / thyroid gland development / Uptake and function of anthrax toxins / positive regulation of protein serine/threonine kinase activity / extrinsic apoptotic signaling pathway via death domain receptors / somatic stem cell population maintenance / positive regulation of peptidyl-serine phosphorylation / MAP kinase kinase kinase activity / protein kinase activator activity / type II interferon-mediated signaling pathway / Schwann cell development / response to axon injury / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors / negative regulation of protein-containing complex assembly / keratinocyte differentiation / neuron projection morphogenesis / response to muscle stretch / ERK1 and ERK2 cascade / myelination / protein serine/threonine/tyrosine kinase activity / positive regulation of autophagy / CD209 (DC-SIGN) signaling / insulin-like growth factor receptor signaling pathway / dendrite cytoplasm / response to glucocorticoid / thymus development / adenylate cyclase activator activity / MAP3K8 (TPL2)-dependent MAPK1/3 activation / protein serine/threonine kinase activator activity / Signal transduction by L1 / cell motility / positive regulation of transcription elongation by RNA polymerase II / wound healing / RAF activation / Signaling by high-kinase activity BRAF mutants / MAP2K and MAPK activation / small GTPase binding / Stimuli-sensing channels / chemotaxis / neuron differentiation / Negative regulation of MAPK pathway / Signaling by RAF1 mutants / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF / Signaling downstream of RAS mutants / cellular senescence / Signaling by BRAF and RAF1 fusions / insulin receptor signaling pathway / late endosome / MAPK cascade / heart development / response to oxidative stress / protein tyrosine kinase activity Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
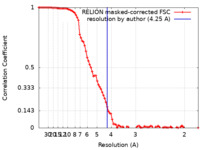
| Method | single particle reconstruction / cryo EM / Resolution: 4.25 Å | |||||||||
 Authors Authors | Dedden D / Ulrich G | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2024 Journal: J Mol Biol / Year: 2024Title: Cryo-EM Structures of CRAF/14-3-3 and CRAF/14-3-3/MEK1 Complexes. Authors: Dirk Dedden / Julius Nitsche / Elisabeth V Schneider / Maren Thomsen / Daniel Schwarz / Birgitta Leuthner / Ulrich Grädler /  Abstract: RAF protein kinases are essential effectors in the MAPK pathway and are important cancer drug targets. Structural understanding of RAF activation is so far based on cryo-electron microscopy (cryo-EM) ...RAF protein kinases are essential effectors in the MAPK pathway and are important cancer drug targets. Structural understanding of RAF activation is so far based on cryo-electron microscopy (cryo-EM) and X-ray structures of BRAF in different conformational states as inactive or active complexes with KRAS, 14-3-3 and MEK1. In this study, we have solved the first cryo-EM structures of CRAF/14-3-3 at 3.4 Å resolution and CRAF/14-3-3/MEK1 at 4.2 Å resolution using CRAF kinase domain expressed as constitutively active Y340D/Y341D mutant in insect cells. The overall architecture of our CRAF/14-3-3 and CRAF/14-3-3/MEK1 cryo-EM structures is highly similar to corresponding BRAF structures in complex with 14-3-3 or 14-3-3/MEK1 and represent the activated dimeric RAF conformation. Our CRAF cryo-EM structures provide additional insights into structural understanding of the activated CRAF/14-3-3/MEK1 complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16660.map.gz emd_16660.map.gz | 55.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16660-v30.xml emd-16660-v30.xml emd-16660.xml emd-16660.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16660_fsc.xml emd_16660_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_16660.png emd_16660.png | 65.2 KB | ||
| Filedesc metadata |  emd-16660.cif.gz emd-16660.cif.gz | 6.9 KB | ||
| Others |  emd_16660_half_map_1.map.gz emd_16660_half_map_1.map.gz emd_16660_half_map_2.map.gz emd_16660_half_map_2.map.gz | 48.7 MB 48.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16660 http://ftp.pdbj.org/pub/emdb/structures/EMD-16660 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16660 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16660 | HTTPS FTP |
-Related structure data
| Related structure data |  8chfMC  8cpdC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16660.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16660.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.91366 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_16660_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_16660_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of dimer phosphorylated C-raf kinase domain with 14-3-3 d...
| Entire | Name: Complex of dimer phosphorylated C-raf kinase domain with 14-3-3 dimer and 2 subunits of Mek1 containing ligand GDC-0623 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of dimer phosphorylated C-raf kinase domain with 14-3-3 d...
| Supramolecule | Name: Complex of dimer phosphorylated C-raf kinase domain with 14-3-3 dimer and 2 subunits of Mek1 containing ligand GDC-0623 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: RAF proto-oncogene serine/threonine-protein kinase
| Macromolecule | Name: RAF proto-oncogene serine/threonine-protein kinase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 73.184477 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEHIQGAWKT ISNGFGFKDA VFDGSSCISP TIVQQFGYQR RASDDGKLTD PSKTSNTIRV FLPNKQRTVV NVRNGMSLHD CLMKALKVR GLQPECCAVF RLLHEHKGKK ARLDWNTDAA SLIGEELQVD FLDHVPLTTH NFARKTFLKL AFCDICQKFL L NGFRCQTC ...String: MEHIQGAWKT ISNGFGFKDA VFDGSSCISP TIVQQFGYQR RASDDGKLTD PSKTSNTIRV FLPNKQRTVV NVRNGMSLHD CLMKALKVR GLQPECCAVF RLLHEHKGKK ARLDWNTDAA SLIGEELQVD FLDHVPLTTH NFARKTFLKL AFCDICQKFL L NGFRCQTC GYKFHEHCST KVPTMCVDWS NIRQLLLFPN STIGDSGVPA LPSLTMRRMR ESVSRMPVSS QHRYSTPHAF TF NTSSPSS EGSLSQRQRS TSTPNVHMVS TTLPVDSRMI EDAIRSHSES ASPSALSSSP NNLSPTGWSQ PKTPVPAQRE RAP VSGTQE KNKIRPRGQR DSSYDWEIEA SEVMLSTRIG SGSFGTVYKG KWHGDVAVKI LKVVDPTPEQ FQAFRNEVAV LRKT RHVNI LLFMGYMTKD NLAIVTQWCE GSSLYKHLHV QETKFQMFQL IDIARQTAQG MDYLHAKNII HRDMKSNNIF LHEGL TVKI GDFGLATVKS RWSGSQQVEQ PTGSVLWMAP EVIRMQDNNP FSFQSDVYSY GIVLYELMTG ELPYSHINNR DQIIFM VGR GYASPDLSKL YKNCPKAMKR LVADCVKKVK EERPLFPQIL SSIELLQHSL PKINRSA(SEP)EP SLHRAAHTED INA CTLTTS PRLPVF UniProtKB: RAF proto-oncogene serine/threonine-protein kinase |
-Macromolecule #2: 14-3-3 protein zeta isoform X1
| Macromolecule | Name: 14-3-3 protein zeta isoform X1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.108514 KDa |
| Sequence | String: MSVDKEELVQ RAKLAEQAER YDDMAAAMKE VTETGVELSN EERNLLSVAY KNVVGARRSS WRVISSIEQK TEGSERKQQM AKEYRVKVE KELREICYDV LGLLDKHLIP KASNPESKVF YLKMKGDYYR YLAEVATGET RNSVVEDSQK AYQDAFEISK A KMQPTHPI ...String: MSVDKEELVQ RAKLAEQAER YDDMAAAMKE VTETGVELSN EERNLLSVAY KNVVGARRSS WRVISSIEQK TEGSERKQQM AKEYRVKVE KELREICYDV LGLLDKHLIP KASNPESKVF YLKMKGDYYR YLAEVATGET RNSVVEDSQK AYQDAFEISK A KMQPTHPI RLGLALNFSV FYYEILNSPD KACQLAKQAF DDAIAELDTL NEDSYKDSTL IMQLLRDNLT LWTSDTQGDG DE PAEGGDN UniProtKB: 14-3-3 protein zeta |
-Macromolecule #3: Dual specificity mitogen-activated protein kinase kinase 1
| Macromolecule | Name: Dual specificity mitogen-activated protein kinase kinase 1 type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO / EC number: mitogen-activated protein kinase kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 43.461938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPKKKPTPIQ LNPAPDGSAV NGTSSAETNL EALQKKLEEL ELDEQQRKRL EAFLTQKQKV GELKDDDFEK ISELGAGNGG VVFKVSHKP SGLVMARKLI HLEIKPAIRN QIIRELQVLH ECNSPYIVGF YGAFYSDGEI SICMEHMDGG SLDQVLKKAG R IPEQILGK ...String: MPKKKPTPIQ LNPAPDGSAV NGTSSAETNL EALQKKLEEL ELDEQQRKRL EAFLTQKQKV GELKDDDFEK ISELGAGNGG VVFKVSHKP SGLVMARKLI HLEIKPAIRN QIIRELQVLH ECNSPYIVGF YGAFYSDGEI SICMEHMDGG SLDQVLKKAG R IPEQILGK VSIAVIKGLT YLREKHKIMH RDVKPSNILV NSRGEIKLCD FGVSGQLIDA MANAFVGTRS YMSPERLQGT HY SVQSDIW SMGLSLVEMA VGRYPIPPPD AKELELMFGC QVEGDAAETP PRPRTPGRPL SSYGMDSRPP MAIFELLDYI VNE PPPKLP SGVFSLEFQD FVNKCLIKNP AERADLKQLM VHAFIKRSDA EEVDFAGWLC STIGLNQPST PTHAAGV UniProtKB: Dual specificity mitogen-activated protein kinase kinase 1 |
-Macromolecule #4: 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin...
| Macromolecule | Name: 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol type: ligand / ID: 4 / Number of copies: 2 / Formula: 29L |
|---|---|
| Molecular weight | Theoretical: 334.372 Da |
| Chemical component information | 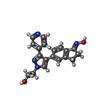 ChemComp-29L: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 2732 / Average exposure time: 7.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 150000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8chf: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)