+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the RESC1-RESC2 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RESC / RNA editing / cryo-EM structure / Trypanosoma brucei / RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmitochondrial mRNA processing / mitochondrial mRNA editing complex / mitochondrial RNA processing / kinetoplast / mRNA modification / mRNA binding / mitochondrion Similarity search - Function | |||||||||
| Biological species |  | |||||||||
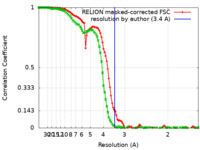
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Dolce LG / Weis F / Kowalinski E | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Structural basis for guide RNA selection by the RESC1-RESC2 complex. Authors: Luciano G Dolce / Yevheniia Nesterenko / Leon Walther / Félix Weis / Eva Kowalinski /   Abstract: Kinetoplastid parasites, such as trypanosomes or leishmania, rely on RNA-templated RNA editing to mature mitochondrial cryptic pre-mRNAs into functional protein-coding transcripts. Processive pan- ...Kinetoplastid parasites, such as trypanosomes or leishmania, rely on RNA-templated RNA editing to mature mitochondrial cryptic pre-mRNAs into functional protein-coding transcripts. Processive pan-editing of multiple editing blocks within a single transcript is dependent on the 20-subunit RNA editing substrate binding complex (RESC) that serves as a platform to orchestrate the interactions between pre-mRNA, guide RNAs (gRNAs), the catalytic RNA editing complex (RECC), and a set of RNA helicases. Due to the lack of molecular structures and biochemical studies with purified components, neither the spacio-temporal interplay of these factors nor the selection mechanism for the different RNA components is understood. Here we report the cryo-EM structure of Trypanosoma brucei RESC1-RESC2, a central hub module of the RESC complex. The structure reveals that RESC1 and RESC2 form an obligatory domain-swapped dimer. Although the tertiary structures of both subunits closely resemble each other, only RESC2 selectively binds 5'-triphosphate-nucleosides, a defining characteristic of gRNAs. We therefore propose RESC2 as the protective 5'-end binding site for gRNAs within the RESC complex. Overall, our structure provides a starting point for the study of the assembly and function of larger RNA-bound kinetoplast RNA editing modules and might aid in the design of anti-parasite drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16592.map.gz emd_16592.map.gz | 80.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16592-v30.xml emd-16592-v30.xml emd-16592.xml emd-16592.xml | 24.7 KB 24.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16592_fsc.xml emd_16592_fsc.xml emd_16592_fsc_2.xml emd_16592_fsc_2.xml | 10.7 KB 13.8 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_16592.png emd_16592.png | 90.8 KB | ||
| Masks |  emd_16592_msk_1.map emd_16592_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16592.cif.gz emd-16592.cif.gz | 6.9 KB | ||
| Others |  emd_16592_additional_1.map.gz emd_16592_additional_1.map.gz emd_16592_additional_2.map.gz emd_16592_additional_2.map.gz emd_16592_half_map_1.map.gz emd_16592_half_map_1.map.gz emd_16592_half_map_2.map.gz emd_16592_half_map_2.map.gz | 91.3 MB 6 MB 80.8 MB 81 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16592 http://ftp.pdbj.org/pub/emdb/structures/EMD-16592 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16592 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16592 | HTTPS FTP |
-Validation report
| Summary document |  emd_16592_validation.pdf.gz emd_16592_validation.pdf.gz | 779.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16592_full_validation.pdf.gz emd_16592_full_validation.pdf.gz | 779 KB | Display | |
| Data in XML |  emd_16592_validation.xml.gz emd_16592_validation.xml.gz | 17.6 KB | Display | |
| Data in CIF |  emd_16592_validation.cif.gz emd_16592_validation.cif.gz | 23.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16592 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16592 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16592 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16592 | HTTPS FTP |
-Related structure data
| Related structure data |  8cdpMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16592.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16592.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||
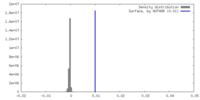
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16592_msk_1.map emd_16592_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
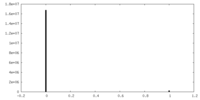
| Projections & Slices |
| ||||||||||||
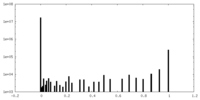
| Density Histograms |
-Additional map: deepEMhancer post process
| File | emd_16592_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepEMhancer post process | ||||||||||||
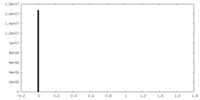
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: relion post process
| File | emd_16592_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | relion post process | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16592_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_16592_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RESC1-RESC2 complex
| Entire | Name: RESC1-RESC2 complex |
|---|---|
| Components |
|
-Supramolecule #1: RESC1-RESC2 complex
| Supramolecule | Name: RESC1-RESC2 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Guide_RNA_associated_protein_-_putative
| Macromolecule | Name: Guide_RNA_associated_protein_-_putative / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.724535 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKHHHHHHSA GLEVLFQGPD SMQNQGSVSQ GALNMRDQQA AAAENVTPER VWALWNEGNL FSLSLAQLQG FLSRCGVRTD PAAKKAAVV RQVEEYLHSK DTTVKGGGQG AASPQQHQQH GQQGGYGRWN QASVMQPETL LDLSQAGFYE GAANMVPKAF Q LLVSDTAP ...String: MKHHHHHHSA GLEVLFQGPD SMQNQGSVSQ GALNMRDQQA AAAENVTPER VWALWNEGNL FSLSLAQLQG FLSRCGVRTD PAAKKAAVV RQVEEYLHSK DTTVKGGGQG AASPQQHQQH GQQGGYGRWN QASVMQPETL LDLSQAGFYE GAANMVPKAF Q LLVSDTAP DVVVSRVNTT AFPGFPSNTE CYTLGASEKD VAIRSRYSKV LQWCCLNMSN LQMDGELYVD FGKLLLKPSV MR KNRRIVS SYTLQQRLQV NHPYTWVPTL PESCLSKIQE QFLQPEGFAP IGKGVQLTYS GTIKRSKDQL HVDLDNKGKV LAV NSAWVN LQTAWCTHAK GPDVRLLLRS RPPIRRQDVE LFASTPIIKL ADDDVADVLP PEHGQLVYLS EDETRLFERV SDRG VTITV REVKRQPLII LRDEEEDPRV EYSLSAHIPA NAAKATDVRA VGLTAFELAG RLAGLVAEDF VREYGCEAKL UniProtKB: Guide RNA associated protein, GAP2 |
-Macromolecule #2: Mitochondrial guide RNA binding complex subunit 2
| Macromolecule | Name: Mitochondrial guide RNA binding complex subunit 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.210234 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MQSFSAAAPA ASGDFSHITR NTVWGLWNEG NLFSLSVPEL AFFLQEHCRV ANVDPRAKKS ALVRQVEEIL SAEQQASATV PQEDNPHAI VVTDYDRAED ALEEADEYGD WGAEPGFEDR RELDFMELSP GRMGERYDPL SPRAFQLLHS ETATDVGIAS I DPSKLPGQ ...String: MQSFSAAAPA ASGDFSHITR NTVWGLWNEG NLFSLSVPEL AFFLQEHCRV ANVDPRAKKS ALVRQVEEIL SAEQQASATV PQEDNPHAI VVTDYDRAED ALEEADEYGD WGAEPGFEDR RELDFMELSP GRMGERYDPL SPRAFQLLHS ETATDVGIAS I DPSKLPGQ SKVKNALAAI HVAPNDANKM RFRMAFEWCL MNIWNMNMPG ELNIGAGKAL YYRSVAKQNR NVMPLWTVQK HL YAQHPYA WFAIASESNV AAMESLAAAL NMSIQQERTT SYKVTIRRMA EFFDCELNGQ LKCTMMNKPW DRFFVSHYIR SKM PDLRYV VRARHPIKKR IADAYLEADI LRSTRDSVQS VLSPELGDVV YCCERVVRKW AKKTATGVTL QLVETKRTPL IITK AGDEG ERLEYEWIVP LPQQAERIDI AALTDELWEY GNKLAAALEE GMEELMVHTM TAVSAY UniProtKB: RESC1/2 CYTH-like domain-containing protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.07 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 63.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)