+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of RESC1-RESC2 bound to gRNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RESC / RNA editing / cryo-EM structure / Trypanosoma brucei / RNA BINDING PROTEIN | |||||||||
| Biological species |  | |||||||||
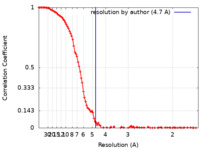
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Dolce LG / Kowalinski E | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Structural basis for guide RNA selection by the RESC1-RESC2 complex. Authors: Luciano G Dolce / Yevheniia Nesterenko / Leon Walther / Félix Weis / Eva Kowalinski /   Abstract: Kinetoplastid parasites, such as trypanosomes or leishmania, rely on RNA-templated RNA editing to mature mitochondrial cryptic pre-mRNAs into functional protein-coding transcripts. Processive pan- ...Kinetoplastid parasites, such as trypanosomes or leishmania, rely on RNA-templated RNA editing to mature mitochondrial cryptic pre-mRNAs into functional protein-coding transcripts. Processive pan-editing of multiple editing blocks within a single transcript is dependent on the 20-subunit RNA editing substrate binding complex (RESC) that serves as a platform to orchestrate the interactions between pre-mRNA, guide RNAs (gRNAs), the catalytic RNA editing complex (RECC), and a set of RNA helicases. Due to the lack of molecular structures and biochemical studies with purified components, neither the spacio-temporal interplay of these factors nor the selection mechanism for the different RNA components is understood. Here we report the cryo-EM structure of Trypanosoma brucei RESC1-RESC2, a central hub module of the RESC complex. The structure reveals that RESC1 and RESC2 form an obligatory domain-swapped dimer. Although the tertiary structures of both subunits closely resemble each other, only RESC2 selectively binds 5'-triphosphate-nucleosides, a defining characteristic of gRNAs. We therefore propose RESC2 as the protective 5'-end binding site for gRNAs within the RESC complex. Overall, our structure provides a starting point for the study of the assembly and function of larger RNA-bound kinetoplast RNA editing modules and might aid in the design of anti-parasite drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16593.map.gz emd_16593.map.gz | 80.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16593-v30.xml emd-16593-v30.xml emd-16593.xml emd-16593.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16593_fsc.xml emd_16593_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_16593.png emd_16593.png | 56.3 KB | ||
| Masks |  emd_16593_msk_1.map emd_16593_msk_1.map | 103 MB |  Mask map Mask map | |
| Others |  emd_16593_additional_1.map.gz emd_16593_additional_1.map.gz emd_16593_additional_2.map.gz emd_16593_additional_2.map.gz emd_16593_half_map_1.map.gz emd_16593_half_map_1.map.gz emd_16593_half_map_2.map.gz emd_16593_half_map_2.map.gz | 7.4 MB 91.3 MB 80.9 MB 80.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16593 http://ftp.pdbj.org/pub/emdb/structures/EMD-16593 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16593 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16593 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16593.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16593.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
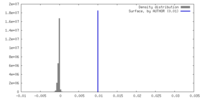
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16593_msk_1.map emd_16593_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
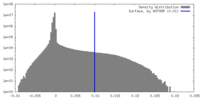
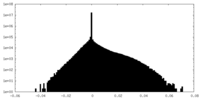
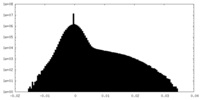
| Density Histograms |
-Additional map: Relion post process
| File | emd_16593_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion post process | ||||||||||||
| Projections & Slices |
| ||||||||||||
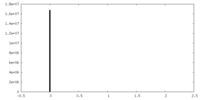
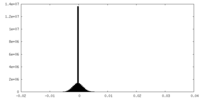
| Density Histograms |
-Additional map: deepEMhancer post process
| File | emd_16593_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | deepEMhancer post process | ||||||||||||
| Projections & Slices |
| ||||||||||||
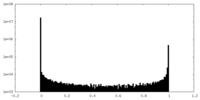
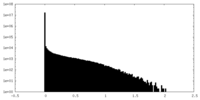
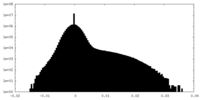
| Density Histograms |
-Half map: #1
| File | emd_16593_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16593_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
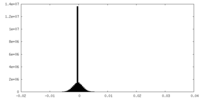
| Density Histograms |
- Sample components
Sample components
-Entire : RESC1-RESC2-gRNA complex
| Entire | Name: RESC1-RESC2-gRNA complex |
|---|---|
| Components |
|
-Supramolecule #1: RESC1-RESC2-gRNA complex
| Supramolecule | Name: RESC1-RESC2-gRNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 118 KDa |
-Macromolecule #1: RESC1
| Macromolecule | Name: RESC1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKHHHHHHSA GLEVLFQGPD SMQNQGSVSQ GALNMRDQQA AAAENVTPER VWALWNEGNL FSLSLAQLQG FLSRCGVRTD PAAKKAAVVR QVEEYLHSKD TTVKGGGQGA ASPQQHQQHG QQGGYGRWNQ ASVMQPETLL DLSQAGFYEG AANMVPKAFQ LLVSDTAPDV ...String: MKHHHHHHSA GLEVLFQGPD SMQNQGSVSQ GALNMRDQQA AAAENVTPER VWALWNEGNL FSLSLAQLQG FLSRCGVRTD PAAKKAAVVR QVEEYLHSKD TTVKGGGQGA ASPQQHQQHG QQGGYGRWNQ ASVMQPETLL DLSQAGFYEG AANMVPKAFQ LLVSDTAPDV VVSRVNTTAF PGFPSNTECY TLGASEKDVA IRSRYSKVLQ WCCLNMSNLQ MDGELYVDFG KLLLKPSVMR KNRRIVSSYT LQQRLQVNHP YTWVPTLPES CLSKIQEQFL QPEGFAPIGK GVQLTYSGTI KRSKDQLHVD LDNKGKVLAV NSAWVNLQTA WCTHAKGPDV RLLLRSRPPI RRQDVELFAS TPIIKLADDD VADVLPPEHG QLVYLSEDET RLFERVSDRG VTITVREVKR QPLIILRDEE EDPRVEYSLS AHIPANAAKA TDVRAVGLTA FELAGRLAGL VAEDFVREYG CEAKL |
-Macromolecule #2: RESC2
| Macromolecule | Name: RESC2 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MQSFSAAAPA ASGDFSHITR NTVWGLWNEG NLFSLSVPEL AFFLQEHCRV ANVDPRAKKS ALVRQVEEIL SAEQQASATV PQEDNPHAIV VTDYDRAEDA LEEADEYGDW GAEPGFEDRR ELDFMELSPG RMGERYDPLS PRAFQLLHSE TATDVGIASI DPSKLPGQSK ...String: MQSFSAAAPA ASGDFSHITR NTVWGLWNEG NLFSLSVPEL AFFLQEHCRV ANVDPRAKKS ALVRQVEEIL SAEQQASATV PQEDNPHAIV VTDYDRAEDA LEEADEYGDW GAEPGFEDRR ELDFMELSPG RMGERYDPLS PRAFQLLHSE TATDVGIASI DPSKLPGQSK VKNALAAIHV APNDANKMRF RMAFEWCLMN IWNMNMPGEL NIGAGKALYY RSVAKQNRNV MPLWTVQKHL YAQHPYAWFA IASESNVAAM ESLAAALNMS IQQERTTSYK VTIRRMAEFF DCELNGQLKC TMMNKPWDRF FVSHYIRSKM PDLRYVVRAR HPIKKRIADA YLEADILRST RDSVQSVLSP ELGDVVYCCE RVVRKWAKKT ATGVTLQLVE TKRTPLIITK AGDEGERLEY EWIVPLPQQA ERIDIAALTD ELWEYGNKLA AALEEGMEEL MVHTMTAVSA Y |
-Macromolecule #3: gRNA gND7(506)
| Macromolecule | Name: gRNA gND7(506) / type: rna / ID: 3 |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: GGGAUAUAAA UACGAUGUAA AUAACCUGUA GUAUAGUUAG UGUAUAUAGU GAAA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.07 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Average electron dose: 62.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)