[English] 日本語
 Yorodumi
Yorodumi- EMDB-16517: Cryo-EM structure NDUFS4 knockout complex I from Mus musculus hea... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure NDUFS4 knockout complex I from Mus musculus heart (Class 2 N-domain). | ||||||||||||
 Map data Map data | globally sharpened focused refinement | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | NADH ubiquinone oxidoreductase / Complex I / OXIDOREDUCTASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationComplex I biogenesis / Respiratory electron transport / blastocyst hatching / Mitochondrial protein degradation / cardiac muscle tissue development / NADH:ubiquinone reductase (H+-translocating) / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone / proton motive force-driven mitochondrial ATP synthesis / respiratory chain complex I ...Complex I biogenesis / Respiratory electron transport / blastocyst hatching / Mitochondrial protein degradation / cardiac muscle tissue development / NADH:ubiquinone reductase (H+-translocating) / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone / proton motive force-driven mitochondrial ATP synthesis / respiratory chain complex I / NADH dehydrogenase (ubiquinone) activity / aerobic respiration / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / NAD binding / FMN binding / nervous system development / myelin sheath / 4 iron, 4 sulfur cluster binding / electron transfer activity / mitochondrial inner membrane / mitochondrion / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
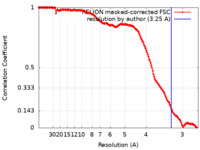
| Method | single particle reconstruction / cryo EM / Resolution: 3.25 Å | ||||||||||||
 Authors Authors | Yin Z / Bridges HR / Agip ANA / Hirst J | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: EMBO J / Year: 2024 Journal: EMBO J / Year: 2024Title: Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 mouse. Authors: Zhan Yin / Ahmed-Noor A Agip / Hannah R Bridges / Judy Hirst /    Abstract: Respiratory complex I (NADH:ubiquinone oxidoreductase) is essential for cellular energy production and NAD homeostasis. Complex I mutations cause neuromuscular, mitochondrial diseases, such as Leigh ...Respiratory complex I (NADH:ubiquinone oxidoreductase) is essential for cellular energy production and NAD homeostasis. Complex I mutations cause neuromuscular, mitochondrial diseases, such as Leigh Syndrome, but their molecular-level consequences remain poorly understood. Here, we use a popular complex I-linked mitochondrial disease model, the ndufs4 mouse, to define the structural, biochemical, and functional consequences of the absence of subunit NDUFS4. Cryo-EM analyses of the complex I from ndufs4 mouse hearts revealed a loose association of the NADH-dehydrogenase module, and discrete classes containing either assembly factor NDUFAF2 or subunit NDUFS6. Subunit NDUFA12, which replaces its paralogue NDUFAF2 in mature complex I, is absent from all classes, compounding the deletion of NDUFS4 and preventing maturation of an NDUFS4-free enzyme. We propose that NDUFAF2 recruits the NADH-dehydrogenase module during assembly of the complex. Taken together, the findings provide new molecular-level understanding of the ndufs4 mouse model and complex I-linked mitochondrial disease. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16517.map.gz emd_16517.map.gz | 322.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16517-v30.xml emd-16517-v30.xml emd-16517.xml emd-16517.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16517_fsc.xml emd_16517_fsc.xml | 15.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_16517.png emd_16517.png | 86 KB | ||
| Masks |  emd_16517_msk_1.map emd_16517_msk_1.map | 347.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16517.cif.gz emd-16517.cif.gz | 6.9 KB | ||
| Others |  emd_16517_half_map_1.map.gz emd_16517_half_map_1.map.gz emd_16517_half_map_2.map.gz emd_16517_half_map_2.map.gz | 323 MB 323 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16517 http://ftp.pdbj.org/pub/emdb/structures/EMD-16517 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16517 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16517 | HTTPS FTP |
-Validation report
| Summary document |  emd_16517_validation.pdf.gz emd_16517_validation.pdf.gz | 1.4 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16517_full_validation.pdf.gz emd_16517_full_validation.pdf.gz | 1.4 MB | Display | |
| Data in XML |  emd_16517_validation.xml.gz emd_16517_validation.xml.gz | 23.4 KB | Display | |
| Data in CIF |  emd_16517_validation.cif.gz emd_16517_validation.cif.gz | 31.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16517 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16517 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16517 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16517 | HTTPS FTP |
-Related structure data
| Related structure data |  8ca4MC  8c2sC  8ca1C  8ca3C  8ca5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16517.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16517.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | globally sharpened focused refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.352 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16517_msk_1.map emd_16517_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_16517_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_16517_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Very long-chain specific acyl-CoA dehydrogenase, mitochondrial
| Entire | Name: Very long-chain specific acyl-CoA dehydrogenase, mitochondrial |
|---|---|
| Components |
|
-Supramolecule #1: Very long-chain specific acyl-CoA dehydrogenase, mitochondrial
| Supramolecule | Name: Very long-chain specific acyl-CoA dehydrogenase, mitochondrial type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial
| Macromolecule | Name: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: NADH:ubiquinone reductase (H+-translocating) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.318336 KDa |
| Sequence | String: MFSLALRARA TGLAAQWGRH ARNLHKTAVH NGAGGALFVH RDTPENNPDT PFDFTPENYK RIEAIVKNYP EGHQAAAVLP VLDLAQRQN GWLPISAMNK VAEVLQVPPM RVYEVATFYT MYNRKPVGKY HIQVCTTTPC MLRDSDSILE TLQRKLGIKV G ETTPDKLF ...String: MFSLALRARA TGLAAQWGRH ARNLHKTAVH NGAGGALFVH RDTPENNPDT PFDFTPENYK RIEAIVKNYP EGHQAAAVLP VLDLAQRQN GWLPISAMNK VAEVLQVPPM RVYEVATFYT MYNRKPVGKY HIQVCTTTPC MLRDSDSILE TLQRKLGIKV G ETTPDKLF TLIEVECLGA CVNAPMVQIN DNYYEDLTPK DIEEIIDELK AGKVPKPGPR SGRFCCEPAG GLTSLTEPPK GP GFGVQAG L UniProtKB: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial |
-Macromolecule #2: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial
| Macromolecule | Name: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: NADH:ubiquinone reductase (H+-translocating) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.904152 KDa |
| Sequence | String: MLAARHFLGG LVPVRVSVRF SSGTTAPKKT SFGSLKDEDR IFTNLYGRHD WRLKGALRRG DWYKTKEILL KGPDWILGEM KTSGLRGRG GAGFPTGLKW SFMNKPSDGR PKYLVVNADE GEPGTCKDRE IMRHDPHKLV EGCLVGGRAM GARAAYIYIR G EFYNEASN ...String: MLAARHFLGG LVPVRVSVRF SSGTTAPKKT SFGSLKDEDR IFTNLYGRHD WRLKGALRRG DWYKTKEILL KGPDWILGEM KTSGLRGRG GAGFPTGLKW SFMNKPSDGR PKYLVVNADE GEPGTCKDRE IMRHDPHKLV EGCLVGGRAM GARAAYIYIR G EFYNEASN LQVAIREAYE AGLIGKNACG SDYDFDVFVV RGAGAYICGE ETALIESIEG KQGKPRLKPP FPADVGVFGC PT TVANVET VAVSPTICRR GGTWFAGFGR ERNSGTKLFN ISGHVNHPCT VEEEMSVPLK ELIEKHAGGV TGGWDNLLAV IPG GSSTPL IPKSVCETVL MDFDALVQAQ TGLGTAAVIV MDRSTDIVKA IARLIEFYKH ESCGQCTPCR EGVDWMNKVM ARFV KGDAR PAEIDSLWEI SKQIEGHTIC ALGDGAAWPV QGLIRHFRPE LEDRMQRFAQ QHRAWQAAS UniProtKB: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial |
-Macromolecule #3: NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial
| Macromolecule | Name: NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: NADH:ubiquinone reductase (H+-translocating) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 79.866688 KDa |
| Sequence | String: MLRIPIKRAL IGLSNSPKGY VRTTGTAASN LIEVFVDGQS VMVEPGTTVL QACEKVGMQI PRFCYHERLS VAGNCRMCLV EIEKAPKVV AACAMPVMKG WNILTNSEKS KKAREGVMEF LLANHPLDCP ICDQGGECDL QDQSMMFGSD RSRFLEGKRA V EDKNIGPL ...String: MLRIPIKRAL IGLSNSPKGY VRTTGTAASN LIEVFVDGQS VMVEPGTTVL QACEKVGMQI PRFCYHERLS VAGNCRMCLV EIEKAPKVV AACAMPVMKG WNILTNSEKS KKAREGVMEF LLANHPLDCP ICDQGGECDL QDQSMMFGSD RSRFLEGKRA V EDKNIGPL VKTIMTRCIQ CTRCIRFASE IAGVDDLGTT GRGNDMQVGT YIEKMFMSEL SGNVIDICPV GALTSKPYAF TA RPWETRK TESIDVMDAV GSNIVVSTRT GEVMRILPRM HEDINEEWIS DKTRFAYDGL KRQRLTEPMV RNEKGLLTYT SWE DALSRV AGMLQNFEGN AVAAIAGGLV DAEALVALKD LLNKVDSDNL CTEEIFPTEG AGTDLRSNYL LNTTIAGVEE ADVV LLVGT NPRFEAPLFN ARIRKSWLHN DLKVALIGSP VDLTYRYDHL GDSPKILQDI ASGRHSFCEV LKDAKKPMVV LGSSA LQRD DGAAILVAVS NMVQKIRVTT GVAAEWKVMN ILHRIASQVA ALDLGYKPGV EAIRKNPPKM LFLLGADGGC ITRQDL PKD CFIVYQGHHG DVGAPMADVI LPGAAYTEKS ATYVNTEGRA QQTKVAVTPP GLAREDWKII RALSEIAGIT LPYDTLD QV RNRLEEVSPN LVRYDDIEET NYFQQASELA KLVNQEVLAD PLVPPQLTIK DFYMTDSISR ASQTMAKCVK AVTEGAQA V EEPSIC UniProtKB: NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial |
-Macromolecule #4: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2
| Macromolecule | Name: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.932675 KDa |
| Sequence | String: MAAAAASRAV GAKLGLREIR VHLCQRSPGS QGVRDFIVQR YVELKKAHPN LPILIRECSE VQPKLWARYA FGQEKTVSLN NLSADEVTR AMQNVLSGKA UniProtKB: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 |
-Macromolecule #5: NADH dehydrogenase [ubiquinone] flavoprotein 3, mitochondrial
| Macromolecule | Name: NADH dehydrogenase [ubiquinone] flavoprotein 3, mitochondrial type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.833504 KDa |
| Sequence | String: MAVSLLLRGG RIRALKAVLL EARVFPGELV SVVRLSTESE KSAKEKELHP KTQSVLKEPE PTDTTTYKNL QHHDYNTYTF LDLNLDLSK FRLPQPSSGR ESPRH UniProtKB: NADH dehydrogenase [ubiquinone] flavoprotein 3, mitochondrial |
-Macromolecule #6: FE2/S2 (INORGANIC) CLUSTER
| Macromolecule | Name: FE2/S2 (INORGANIC) CLUSTER / type: ligand / ID: 6 / Number of copies: 2 / Formula: FES |
|---|---|
| Molecular weight | Theoretical: 175.82 Da |
| Chemical component information |  ChemComp-FES: |
-Macromolecule #7: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 7 / Number of copies: 3 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Macromolecule #8: FLAVIN MONONUCLEOTIDE
| Macromolecule | Name: FLAVIN MONONUCLEOTIDE / type: ligand / ID: 8 / Number of copies: 1 / Formula: FMN |
|---|---|
| Molecular weight | Theoretical: 456.344 Da |
| Chemical component information |  ChemComp-FMN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.82 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.14 Component:
Details: pH was corrected at room temperature ~22 C | |||||||||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Support film - Material: GOLD / Support film - topology: HOLEY | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.9 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)